David McCandlish and Juannan Zhou, both quantitative biologists at the Cold Spring Harbor Laboratory, have designed a new algorithm that has predictive power, providing researchers the ability to observe the way particular genetic mutations combine to make crucial proteins change over the duration of a species’ evolution.
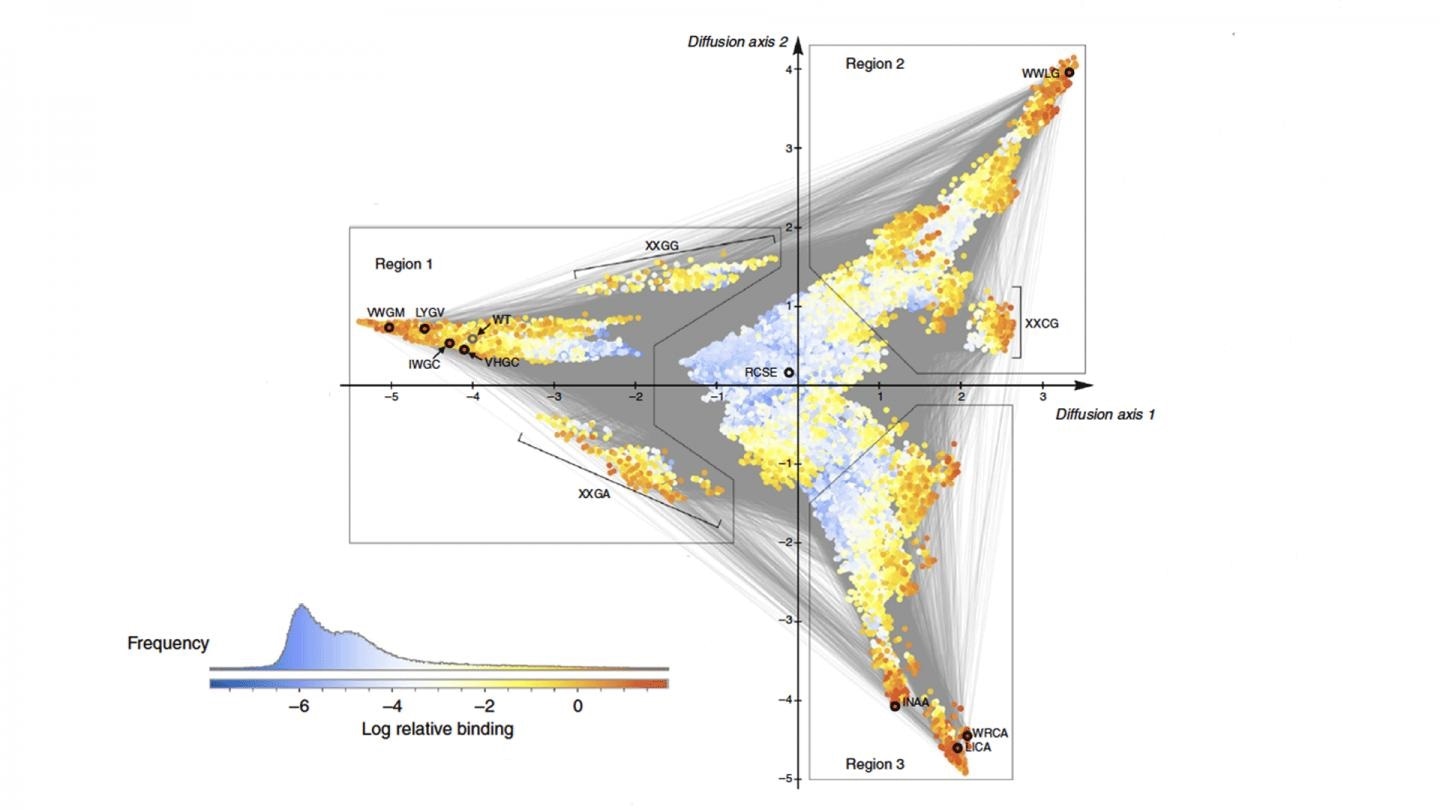
The algorithm called “minimum epistasis interpolation” results in a visualization of how a protein could evolve to either become highly effective or not effective at all. They compared the functionality of thousands of versions of the protein, finding patterns in how mutations cause the protein to evolve from one functional form to another. Image Credit: McCandlish lab/CSHL, 2020.
The new algorithm, known as “minimum epistasis interpolation,” has been detailed in the Nature Communications journal. It helps observe the way a protein is likely to evolve to become extremely effective or not effective at all.
The researchers compared the functionality of scores of protein versions and found patterns in how mutations make the protein to transform from one functional form to another.
“Epistasis” elucidates all kinds of interactions that occur between genetic mutations, where the influence of one gene relies on the presence of another. In a majority of the cases, researchers believed that when reality fails to match with their predictive models, then such gene-to-gene interactions would be involved.
Keeping this aspect in mind, McCandlish designed this novel algorithm assuming that every mutation is significant. The name “Interpolation” defines the act of predicting the mutations’ evolutionary path a species may undergo to get optimal protein function.
The scientists developed the new algorithm by testing the impacts of particular mutations taking place in the genes that constitute the streptococcal GB1 protein.
The team selected the GB1 protein due to its intricate structure that would produce a large number of potential mutations that could be combined in many promising ways.
Because of this complexity, visualization of this data set became so important. We wanted to turn the numbers into a picture so that we can understand better what [the data] is telling us.”
David McCandlish, Cold Spring Harbor Laboratory
The visualization is similar to a topological map. Color and height correspond to the level of protein activity, and the distance between the points on the map indicates the time taken for the mutations to evolve to that degree of activity.
The GB1 protein naturally starts with a modest level of protein activity but can evolve to a level of higher protein activity via a series of mutations that take place at many different places.
McCandlish compares the protein’s evolutionary path to hiking, in which the protein is a hiker attempting to reach the best or highest mountain peaks most efficiently. Genes tend to evolve in the same way with a mutation looking for the path of increased efficiency and the least resistance.
To reach the next best high peak in the mountain range, the hiker will tend to move along the ridgeline rather than hike all the way back down to the valley. Another potentially difficult ascent is also avoided by going along the ridgeline efficiently.
In the visualization, the valley is the blue region, where a mix of mutations leads to the lowest levels of protein activity.
The new algorithm demonstrates the optimal level of each potential mutant sequence and the time taken for a single genetic sequence to mutate into any of the several other potential sequences.
The tool’s predictive power may prove specifically useful in circumstances such as the COVID-19 pandemic. Scientists should be aware of the evolution of a virus to learn where and when to intercept this pathogen before it transforms into its most dangerous form.
McCandlish explained that the new algorithm can even help “understand the genetic routes that a virus might take as it evolves to evade the immune system or gain drug resistance. If we can understand the likely routes, then maybe we can design therapies that can prevent the evolution of resistance or immune evasion.”
A predictive genetic algorithm like this has more potential applications for such, such as agriculture and drug development.
You know, at the very beginning of genetics ... there was all this interesting speculation as to what these genetic spaces would look like if you could actually look at them. Now we’re really doing it! That’s really cool.”
David McCandlish, Cold Spring Harbor Laboratory
Source:
Journal reference:
Zhou, J., et al. (2020) Minimum epistasis interpolation for sequence-function relationships, Nature communications. doi.org/10.1038/s41467-020-15512-5.