There has been very little knowledge of the ways in which transposable elements play a role in gene regulation. Transposable elements are small DNA pieces with the ability to replicate and scatter themselves in the genome.
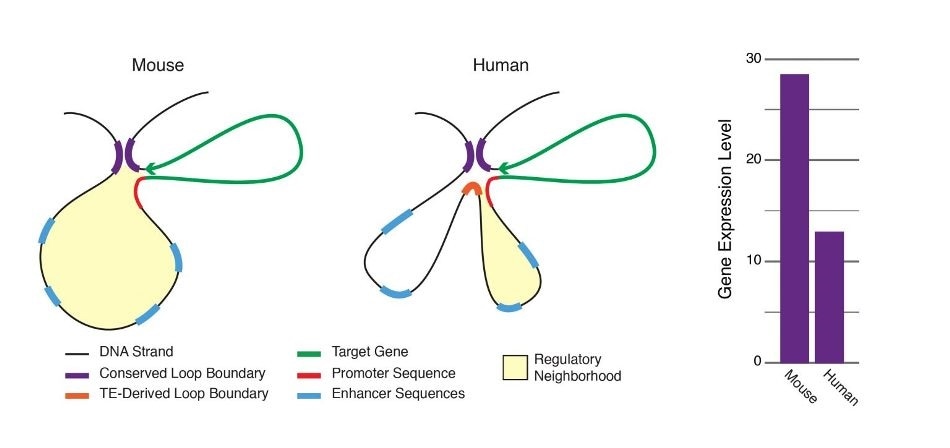
Chromatin loops are important for gene regulation because they define a gene’s regulatory neighborhood, which contains the promoter and enhancer sequences responsible for determining its expression level. Remarkably, transposable elements (TEs) are responsible for creating around 1/3 of all loop boundaries in the human and mouse genomes and contribute up to 75% of loops unique to either species. When a TE creates a human-specific or mouse-specific loop it can change a gene’s regulatory neighborhood, leading to altered gene expression. The illustration shows a hypothetical region of the human and mouse genomes in which four enhancer sequences for the same target gene fall within a conserved loop. In this example, a TE-derived loop boundary in the human genome (orange bar) shrinks the regulatory neighborhood, preventing two of four enhancers from interacting with their target gene’s promoter sequence. The net result is reduced gene expression in human relative to mouse. Looping variations such as these appear to be an important underlying cause of differential gene regulation across species and between different human cell types, suggesting that TE activity may play significant roles in evolution and disease. Image Credit: Adam Diehl.
Despite constituting almost half of the human genome, these were usually ignored and commonly considered as “useless junk,” with very little role, if any, in cellular activity.
New research by Adam Diehl, Ningxin Ouyang, and Alan Boyle from the University of Michigan Medical School and members of the U-M Center for RNA Biomedicine reveals that transposable elements have a key role in controlling gene expression with implications to offer better insights into genetic evolution.
Transposable elements tend to move around the cell, and in contrast to earlier predictions, the study authors discovered that when they move to various sites, they sometimes alter the way DNA strands interact in 3D space, and thus the 3D genome structure.
It seems one-third of the 3D contacts in the genome, in fact, emerge from transposable elements, which results in an oversized contribution by these regions to looping variation and reveals their highly crucial role in genetic expression and evolution.
A protein named CTCF is the main component that governs the 3D structure. The focus of this study was specifically on how transposable elements form new CTCF sites that, alternatively, hijack current genomic structure to create new 3D contacts in the genome.
The researchers demonstrate that these usually form variable loops that can affect the regulatory activity and gene expression in the cell.
These outcomes were seen in mouse cells and human cells and reveal how transposable elements bring about interspecies divergence and intraspecies variation. The findings will pave the way for further studies in areas such as regulatory evolution, gene regulation, transposable element biology, and looping divergence.
To simplify this study, the researchers created software called MapGL to track the physical gain and loss of short genetic sequences among species. For instance, a sequence that occurred in the most common ancestor could have been lost somewhere or, by contrast, could have been missing in the common ancestor but later emerged in the human genome.
MapGL facilitates making predictions in relation to the evolutionary impacts of structural variations between species and renders analyses of this type more accessible.
For this study, their input was a set of CTCF binding sites labeled by MapGL to demonstrate that a sequence gain/loss process accounts for several differences in CTCF binding between mice and humans.
With a background in molecular biology and computer science, Alan Boyle said that he has always been intrigued by gene regulation.
It’s like a complex circuit: perturbing gene regulation through changes to the three-dimensional structure of the genome can have very different and wide-ranging outcomes.”
Alan Boyle, Assistant Professor, Department of Computational Medicine and Bioinformatics, Department of Human Genetics, University of Michigan Medical School
For Adam Diehl, this study is a follow-up of the major findings that started in the late 1800s, when researchers first observed the shape of chromosomes through microscopes. They looked at the differences in shape between cells and observed that the shape within the nuclei remained unaltered between daughter and mother cells.
After several decades, transposable elements were found at Cornell University, his alma mater—jumping genes could alter the phenotypes of corn plants.
In the 1970s, the focus of researchers turned to ways in which the genes were used, since the genes between chimpanzees and humans are way too similar to account for the differences between the species.
It’s so exciting to be able to synthesize all this knowledge, and contribute to the next step of the story of species evolution.”
Alan Boyle, Assistant Professor, Department of Computational Medicine and Bioinformatics, Department of Human Genetics, University of Michigan Medical School
The researchers will further analyze the effect of transposable elements on the 3D genome. However, the focus now will be on a single human population sample rather than between species.
Further studies will be to perform experimental follow-up with a new sequencing technique that can identify transposable element insertions that differ across human populations.
This technique was created in partnership with Ryan Mills’s lab, at the University of Michigan Medical School. It is predicted that further results will advance the insights into the regulatory role of the transposable elements with potential applications to neurodegenerative diseases.
Source:
Journal reference:
Diehl, A. G., et al. (2020) Transposable elements contribute to cell and species-specific chromatin looping and gene regulation in mammalian genomes. Nature Communications. doi.org/10.1038/s41467-020-15520-5.