Complex organisms experience growth, diseases, and death that all depend on the flow of information, starting right from genes in DNA, followed by the transcription of these genes into RNA, and subsequently the translation of that transcript into proteins, which are consequently responsible for building most of the living organism.
Favor or optimal codons comparing to nonoptimal codons. Image Credit: Toshifumi Inada, Tohoku University.
Proteins that regulate this entire process are also subject to this overarching flow of information for their survival.
Now, scientists have identified an earlier unknown function of a protein group known as the Ccr4-Not complex that may offer a deeper understanding of the development of various diseases, including Alzheimer’s.
The results of the study were published in the Science journal on April 17th, 2020.
The Ccr4-Not complex is involved in so many aspects of gene expression that we might as well call it the ‘Swiss Army Knife’ of protein production.”
Toshifumi Inada, Study Lead and Professor, Graduate School of Pharmaceutical Sciences, Tohoku University
The best interpreted aspect of the role of the Ccr4-Not complex is that it is implicated in the destruction of the messenger RNA or mRNA.
The molecules of mRNA are similar to instruction manuals that inform the ribosomes, the protein-making machinery of the cells, on how to build the proteins.
The number of proteins translated by the ribosomes is very important, and so is the rate of that translation. Individuals would not prefer too fast or too slow, or too many or too few. Consequently, the concentrations of these proteins rely on the quantity of mRNA.
Therefore, the regulation of mRNA destruction, particularly when these instruction manuals have errors in them, is crucial to control protein production.
But until now, how Ccr4-Not did this has remained elusive.”
Toshifumi Inada, Study Lead and Professor, Graduate School of Pharmaceutical Sciences, Tohoku University
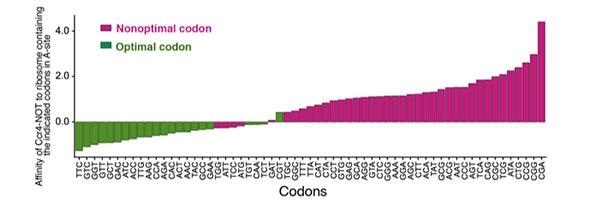
The instruction manuals relating to the mRNAs are made up of various three-unit codes known as “codons” that instruct the ribosomes for the types of amino acids to be used. Incidentally, amino acids are the building blocks of proteins.
However, even with this kind of error-tolerant redundancy, a preference, or to utilize the formal word, bias, would still be there toward a specific synonym codon since it accelerates the process. For some reason, if an instruction manual lacks this codon bias, the system would not work at the pace it should.
The scientists discovered that the Ccr4-Not complex is constantly looking for slow-poke mRNA that lacks this codon bias. They also found that, when this complex detects a slow-poke mRNA, a portion of it binds itself to the ribosome, which consequently causes the degradation of the faulty mRNA.
However, if the Ccr4-Not complex itself happens to be defective, that is, missing the portion that would bind to the ribosome that reads the defective mRNA, the Ccr4-Not complex will lose this critical capacity to detect and destroy these faulty mRNAs that produce the proteins at the wrong pace.
The production of proteins that takes place at the wrong pace can lead to incorrect concentration, shape, or location of proteins, all of which have been linked to a wide range of diseases such as Alzheimer’s and Huntington’s.
The latest study provides an understanding of how the abnormalities of the Ccr4-Not complex play a role in such diseases through inadequate regulation of the speed of protein production.
Currently, the scientists are planning to study whether the management of this speed of a normal working Ccr4-Not complex would also make it possible to regulate protein folding and transport of the proteins to their suitable destination in the body.
Source:
Journal reference:
Buschauer, R., et al. (2020) The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science. doi.org/10.1126/science.aay6912.