InsideOutBio expert Dr Alan Herbert has explained a pioneering research in a paper recently published online by the Royal Society Open Science journal.
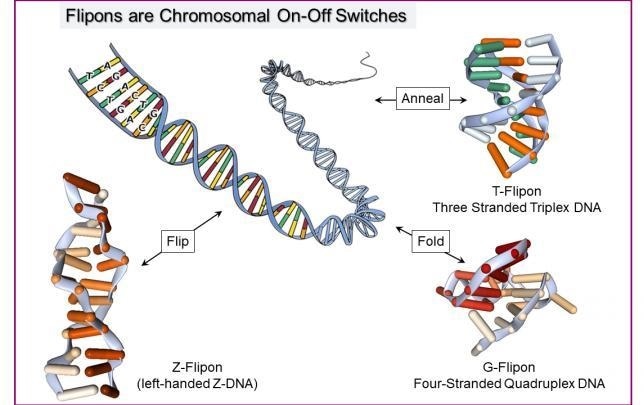
Flipons are scattered through the genome. They switch between the regular B-conformation and an alternate DNA structure. There are different classes of flipons with different numbers of DNA strands (2 for Z-DNA, 3 for a triplex and 4 for a quadruplex) and DNA sequence requirements. Flipons alter how a chromosome is read out. The message produced depends on the flipon conformation. They act as digital switches to turn one message on or another message off. Image Credit: Alan Herbert.
The study targets the digital genome that makes use of programmable DNA-based on-off switches to alter the readout of genetic data.
The genome’s digital rewiring involves switch elements known as flipons. Flipons accelerate the emergence of multi-cellular organisms. The flipon approach is considered a less risky evolution depending only on mutations.
Previously coding by DNA was described as analog. Going digital has greatly increased the genome's storage capacity. Different programs can be run by compiling the information differently.”
Dr Alan Herbert, InsideOutBio
Flipons are essentially DNA sequences that have the ability to adopt different types of DNA conformations. They behave as on-off switches, with every switch modifying the program read out from the DNA code. Changes occur in flipon settings based on the context.
Each setting results in a different set of instructions that have to be followed by the cell. DNA damage repair pathways and innate immune responses provide examples of the behavior of flipons. Flipon conformation establishes whether these repair pathways are active or not.
The evolution of multi-cellular organisms is accelerated by flipons, which spread through the genome via a copy-and-paste mechanism. The flipons function as an on-off switch to modify the way cells gather their operating instructions. Flipons can also be programmed.
Organisms can improve the flipon settings by learning. Survival is improved in clades that learn best. These clades acclimatize and reproduce faster when compared to their competition.
Different categories of flipon exist: Z-flipons can create left-handed DNA by turning over the bases in right-handed DNA. The switch occurs in active genes and helps localize RNA processing complexes to that area.
T-flipons contain three strands to create triplexes. The RNAs required to program a specific result are localized by these flipons. G-flipons are essentially four-stranded structures that trigger repair programs when DNA is damaged.
Flipons help compile several different messages from a solitary genomic sequence. They create more diversity than is possible by rearrangements of DNA or by mutation. These flipons build on already successful adaptations and do not damage them. The recently compiled transcripts function with the old to improve survival.
Many different ways can be used to program the flipons. Modifications of DNA affect how they flip easily from an off-state to an on-state. Proteins also control flipon conformation but the programming needs work. Energy is exchanged for information.
Such a trade-off creates a bigger message space to examine and exploit. When flipons freeze in one state or the other, it usually leads to a disease. A few examples of Mendelian disease caused by flipons have been provided in the paper.
The flipon approach is believed to be less risky when compared to other forms of evolution. Earlier studies focused on DNA mutation as the main driver of change. Mutations alter the sequence of DNA that codes for a protein. This is a random process and cannot be reversed easily.
On the other hand, flipons are both reversible and programmable. They only modify the way messages are compiled from DNA but do not modify the DNA coding sequence. They create variability without the risks linked to mutation. Natural selection relies on variability to identify the most optimal method for the survival and reproduction of an organism.
The paper targets the role of ALU repeat elements in the genome’s digital rewiring. Such elements contain around 11% of the human genome. They proliferate by a copy-and-paste mechanism that relies on reverse transcription of their RNA and DNA. They are believed to be responsible for some of the variations between apes and humans. These ALU elements, once considered the invaders, currently improve the evolution of their host.
Source:
Journal reference:
Herbert, A., (2020) ALU non-B-DNA conformations, flipons, binary codes and evolution. Royal Society Open Science. doi.org/10.1098/rsos.200222.