Cold Spring Harbor Laboratory (CSHL) has developed a novel method that employs DNA sequencing to efficiently plot long-range connections between various brain regions.
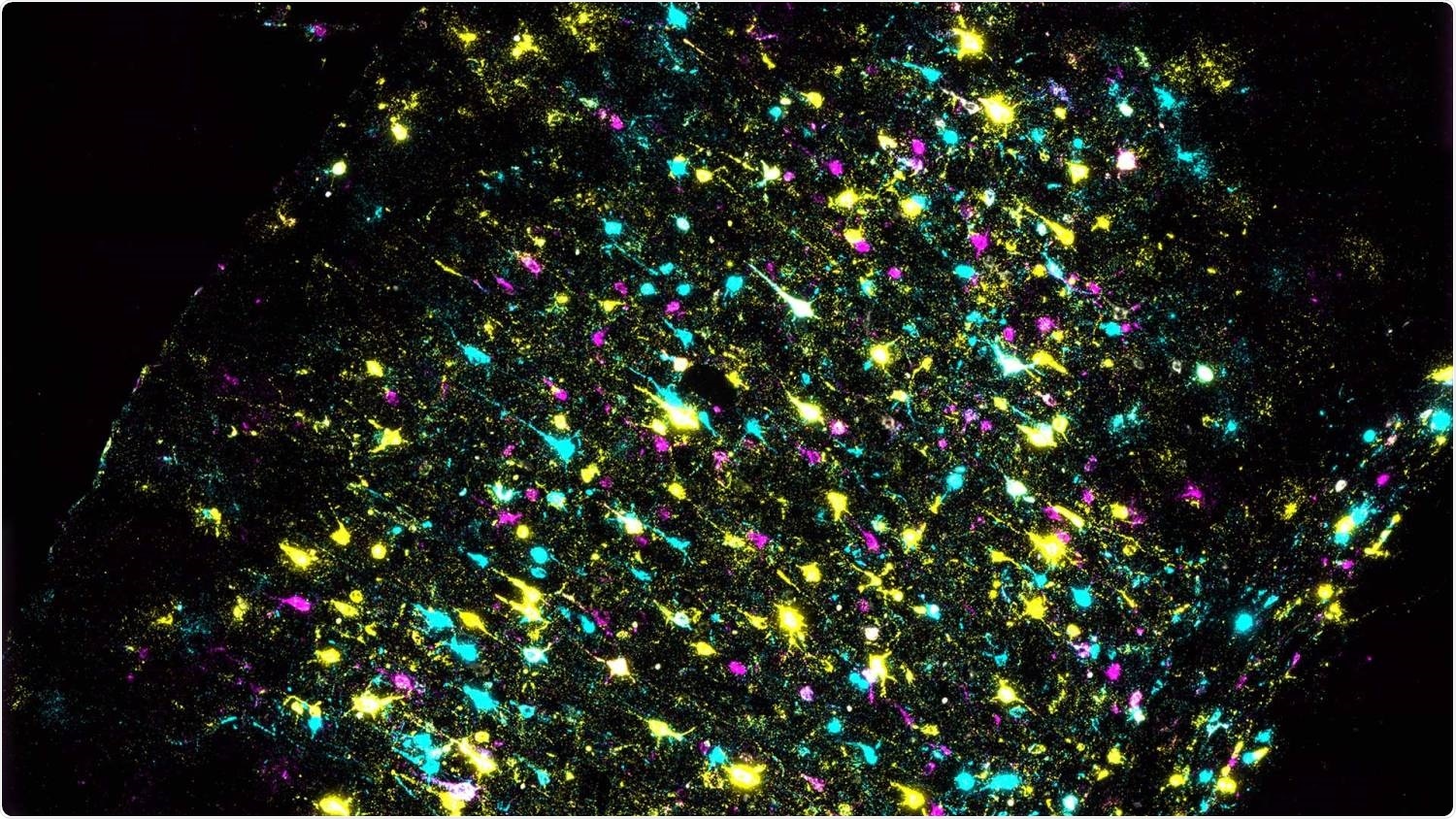
A section of mouse brain stained to identify individual neurons and all their connections. Each color represents a different DNA barcode. Image Credit: Xiaoyin Chen, Zador lab/Cold Spring Harbor Laboratory.
The method significantly decreases the cost of plotting brain-wide connections when compared to conventional microscopy-based techniques.
Anatomical maps are required by neuroscientists to figure out how data flows from one area of the brain to another.
Charting the cellular connections between different parts of the brain—the connectome—can help reveal how the nervous system processes information, as well as how faulty wiring contributes to mental illness and other disorders.”
Longwen Huang, Postdoctoral Researcher, Cold Spring Harbor Laboratory
Huang works in Anthony Zador’s laboratory.
It is a costly and time-consuming process to create these maps and demands enormous efforts that are beyond the reach of a majority of the scientific communities.
Scientists often use fluorescent labels to track the paths of neurons. These fluorescent labels can emphasize how each cell branches through a twisted neural network to locate and attach with its targets.
However, the palette of fluorescent labels appropriate for this study is rather restricted. Scientists can introduce different colors of dyes into two or three regions of the brain and then track the connections emerging from those regions.
This procedure can be repeated to target new areas to view more connections. Moreover, this process should be done scores of times to create a brain-wide map, and each time new research animals are used.
The technique, known as brain-wide individual animal connectome sequencing (BRICseq) and developed in the Zador laboratory, adopts a different method.
We no longer label brain regions and their projections using colors. We are labeling them using nucleotide sequences.”
Longwen Huang, Postdoctoral Researcher, Cold Spring Harbor Laboratory
When the four letters of the DNA code are combined into short “barcodes,” an almost infinite number of labels is generated that can differentiate one cell from the other. Post labeling, the scientists employed DNA sequencing to examine small segments of brain tissues, decoding every recurring barcode as a signal of a cellular connection.
The diversity of barcodes is really high compared to the number of colors we can use in science research. So now we can really label a huge amount of neurons and brain regions per animal, which allows us to map projections from multiple brain regions using these barcodes.”
Longwen Huang, Postdoctoral Researcher, Cold Spring Harbor Laboratory
The researchers, including Justus Kebschull, a former graduate student who collaborated with Huang to design the method, have reported in the Cell journal that the BRICseq method precisely plots region-to-region connectivity in the mice brain. They stated that the method can be extensively accessed and adapted to other kinds of organisms.
Mouse brain connectome
This animation shows how mouse brain cells connect to one another—a sort of wiring diagram of the brain. Researchers in Anthony Zador’s lab at Cold Spring Harbor Laboratory have developed a technique to map many of these connections at once using a “DNA barcode”—a battery of short DNA snippets. Other techniques to determine how cells connect are far more laborious and expensive. Video Credit: Cold Spring Harbor Laboratory.
Source:
Journal reference:
Huang, L., et al. (2020) BRICseq Bridges Brain-wide Interregional Connectivity to Neural Activity and Gene Expression in Single Animals. Cell. doi.org/10.1016/j.cell.2020.05.029.