A group of biomolecular engineers from Rice University has now identified a C-worthy method that significantly improves the precision of gene editing.
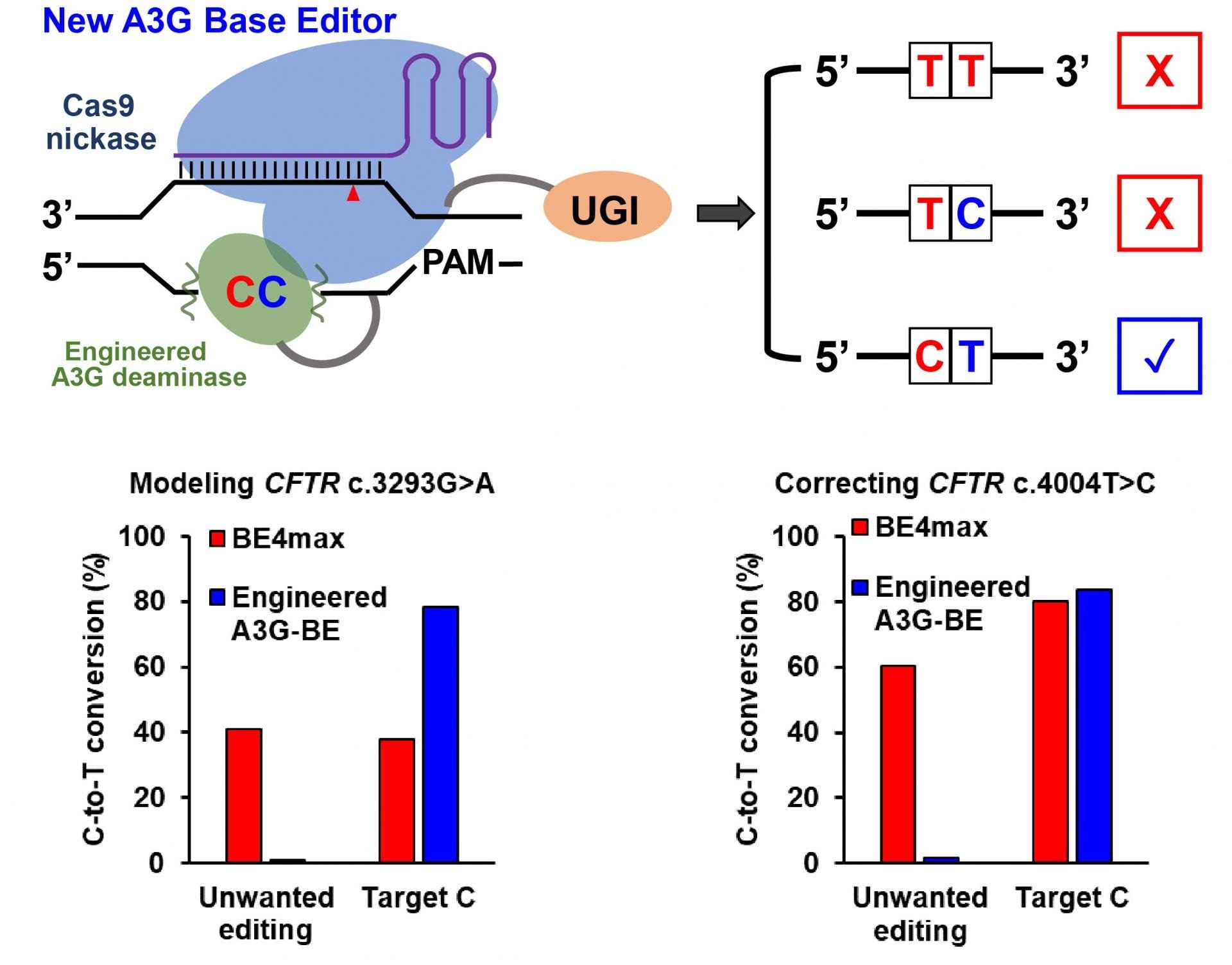
When consecutive cytosines are positioned in the editing window, the new A3G base editor developed at Rice University precisely modifies the single targeted C while minimizing unwanted C editing. Image Credit: Gao Lab/Rice University.
The laboratory of biomolecular engineer Xue Sherry Gao from Rice University has introduced a range of tools that boosts the precision of CRISPR-based edits in disease sequence models by up to 6000 times when compared to that of BE4max, a present-day base editor that is believed to be sophisticated.
The study has been published in Science Advances, an open-access journal.
Cytosine base editors are capable of converting cytosines (C) to thymines (T) in the human genome, which contains as many as three billion As (adenine), Gs (guanine), Ts, and Cs. The base pairs of A-T and C-G encode the genetic data in DNA. Even if a single inaccurate base is present in the human genome—a mutation—it can result in genetic diseases.
T-to-C mutations called single nucleotide polymorphisms account for somewhere around 38% of human pathogenic diseases. Cytosine base editors provide great promise to potentially treat these diseases by reversing the C mutation back to T.”
Xue Sherry Gao, Study Corresponding Author, Department of Bioengineering, Rice University
Gao added, “However, when there is a ‘bystander’ C located right upstream of the targeted C, the previous technology could not distinguish between the Cs, and both would be changed to Ts. We really only want to correct the disease-relevant C to a T and leave the bystander C unmodified.”
That provided the motivation for this project. We want to engineer a new cytosine base editor that can precisely modify the single targeted C while minimizing the unwanted C editing when consecutive ‘CCs’ are positioned in the editing window.”
Xue Sherry Gao, Study Corresponding Author, Department of Bioengineering, Rice University
The Gao laboratory is aiming to develop base editors via a range of protein-engineering efforts. The novel cytosine base editors, known as A3G-BEs, have significantly increased the accuracy by simply editing the second of consecutive Cs.
To place their examinations in “disease-relevant contexts,” the researchers from Gao laboratory employed their tools to alter human cells and thus produce cystic fibrosis as well as many other disease model cell lines.
All the cell lines demonstrated considerable success at accurately producing the required pathogenic C-to-T mutation, specifically the cystic fibrosis cells, in which all the three A3G-BE variants ideally altered over 50% of the time when compared to 0.6% for the BE4max base editor.
In addition, the Gao laboratory validated the potential of its novel A3G-Bes to rectify mutations in disease treatment applications, such as holocarboxylase synthetase deficiency, cystic fibrosis, and pyropoikilocytosis—a kind of anemia.
The A3G-BEs greatly exceeded the BE4max base editor in experiments performed on cell models comprising pathogenetic mutations. With regard to holocarboxylase synthetase deficiency, the editor optimally rectified only the target C nucleotides in over 50% of the sequences, with a correction that is 6,496 times higher than that of the BE4max base editor.
We also identified 540 human pathogenic single nucleotide polymorphisms that could be precisely correctable by our A3G-Bes. A3G-BE also appears to decrease off-target edits (unwanted edits to other parts of the genome that could introduce mutations) at both the DNA and RNA levels.”
Xue Sherry Gao, Study Corresponding Author, Department of Bioengineering, Rice University
Reducing off-targets has been a major objective of CRISPR research.
“There are three billion base pairs in humans. I believe this technology’s level of precision is going to be a significant contributor toward treating genetic disease,” concluded Gao.
Source:
Journal reference:
Lee, S., et al. (2020) Single C-to-T substitution using engineered APOBEC3G-nCas9 base editors with minimum genome- and transcriptome-wide off-target effects. Science Advances. doi.org/10.1126/sciadv.aba1773.