At the University California San Diego, bioengineers have demonstrated that RNA produced by the human genome is found on the surface of human cells, indicating a more comprehensive role for RNA in cell-to-environment and cell-to-cell interactions than previously believed.
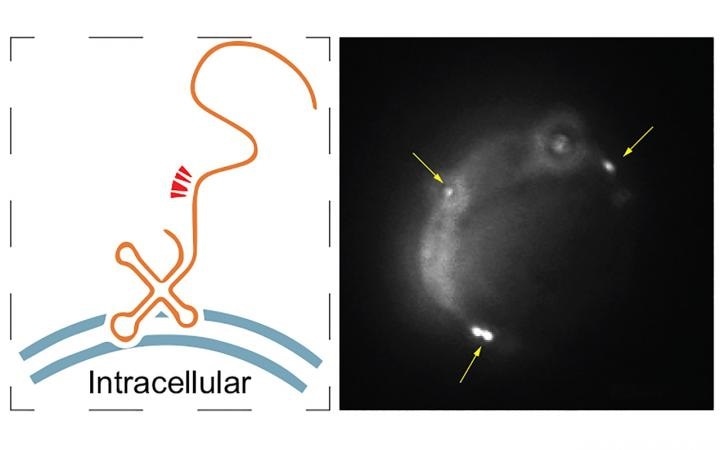
(Left) A hypothetical model of the relative positions of FISH probes (red arrowheads) on a membrane-bound RNA fragment. (Right) A single molecule RNA fluorescence in situ hybridization image of maxRNAs (yellow arrows). Image Credit: Zhong Lab.
Human cells, which are not undergoing cell death, contain this new kind of membrane-associated extracellular RNA, or maxRNA. This provides a better understanding of the contribution of nucleic acids—specifically RNA—to the functions of cell surface.
The maxRNAs as well as the molecular technologies designed to examine the cell surface to detect these maxRNAs have been described in a study published in the Genome Biology journal on September 10th, 2020.
The cell’s surface is to a cell like the face is to a person. It is the most important part for recognizing what type of cell it is, for example a good actor—like a T cell—or a bad actor like a tumor cell—and it aids in communication and interactions.”
Sheng Zhong, Study Corresponding Author and Bioengineering Professor, Jacobs School of Engineering, University California San Diego
Although scientists are considerably aware of other components of the surface of a cell, such as lipids, glycans, and proteins, they do not know much about RNA; excluding a few factors, the RNA created by the human nuclear genome was not believed to occur on the surface of human cells with undamaged cell membranes.
The finding that RNA indeed occurs naturally as a cell surface molecule could play a crucial role in gaining a deeper insight into the genome and designing more effective therapeutics.
This discovery expands our ability to interpret the human genome, because we now know a portion of the human genome may also regulate how a cell presents itself and interacts with other cells through the production of maxRNA.”
Norman Huang, Study First Author and Bioengineering PhD Student, University California San Diego
A deeper interpretation of the maxRNA may also lead to novel strategies for therapeutics development. Furthermore, since the maxRNA is found on the outside surface of the cell, it is easier for therapeutics to reach, and added to this, RNA can be targeted by certain antisense oligonucleotides, which are relatively easier to create when compared to other agents, like antibodies.
To test the presence of RNA on the surface of human and mouse cells, bioengineers in Zhong’s laboratory developed a nanotechnology known as Surface-seq.
The team based this nanotechnology on a method employed by Liangfang Zhang, a Professor from the University California San Diego, to produce very small nanosponges enclosed in natural cell membranes—a process in which the plasma membrane is extracted from cells and assembled around polymeric cores.
This process keeps the surface molecules on the membrane facing outwards and thus sustains the right-side-out orientation of the cell membrane. The stable coating onto the polymeric core and the process of cell membrane purification guarantee the removal of intracellular contents, enabling scientists to spot RNA that is stably related to the extracellular layer of the cell membrane.
The team subsequently defined the functional attributes, cell-type specificity, and sequences of these maxRNA molecules. These molecules were utilized as the input of the construction and sequencing of Surface-seq library.
Apart from associating with Zhang, Zhong also teamed up with professor Zhen Chen’s laboratory in the Beckman Research Institute at City of Hope.
The joint research team intends to additionally explore how the maxRNA is carried to the surface of the cells and affixed there. The team has also planned to further analyze the diversity of genes, cell types, biogenesis pathways, and environmental cues for the expression of maxRNAs and the role played by them in cellular functions.
Source:
Journal reference:
Huang, N., et al. (2020) Natural display of nuclear-encoded RNA on the cell surface and its impact on cell interaction. Genome Biology. doi.org/10.1186/s13059-020-02145-6.