In this interview, Dr. Aimee Murray speaks to AZoLifeSciences about her latest research that looks at a way of reducing antibiotic resistance build-up in waterways.
Increasing antibiotic resistance is a global health problem. What provoked your research into antibiotic resistance?
I have always been interested in microbiology and antibiotic resistance was one of the most obvious ways microbiology relates to real life. I did Biology as an undergraduate but focused on microbiology wherever I could and I really enjoyed learning about resistance. I even did my undergraduate dissertation on MRSA (methicillin-resistant Staphylococcus aureus), which got me interested in doing research. I then did my Ph.D. on antibiotic resistance in the environment.
When I was doing the background reading for my interview, it all made so much sense and seemed so obvious and important that I could not believe we had not been taught anything about resistance in the environment, and I just wanted to learn more.
Image Credit: Kateryna Kon/Shutterstock.com
How do bacteria become resistant to antibiotics?
There are a couple of ways. Bacteria can become resistant through mutations in their genome. For example, if the way an antibiotic works is by binding to a protein and a mutation occurs in that protein that means the antibiotic can no longer bind to it, that bacterium is now resistant.
The other way bacteria can become resistant to antibiotics is through horizontal gene transfer, where entire resistance genes are acquired by bacteria or shared between bacteria. Mutation and horizontal gene transfer can both be caused by antibiotics and many other things.
Roughly 70% of the antibiotics we take end up in the natural environment. How does this occur?
When we take antibiotics, they are not fully broken down in our body. That is actually a good thing, because if we break them down too quickly then they will not be at high enough levels to do their job (kill or inhibit the bacteria in our body that are causing our infection). As a result, we excrete some active antibiotics in our urine and feces, which end up in wastewater treatment plants.
Wastewater treatment plants are effective at removing most bacteria, but they are not designed to remove antibiotics, so some are not fully broken down during the wastewater treatment process. Antibiotics are then released into the aquatic environment in wastewater effluent (the liquid part of treated human waste).
There are also other ways antibiotics can end up in the environment. For example, runoff from farms spread with human sewage sludge (the solid part of treated human waste) or animal manure, as antibiotics are also used a lot in agriculture. Antibiotic manufacturing plants can also be a major source of antibiotics entering the environment.

Image Credit: Daniel Jedzura/Shutterstock.com
Wastewater systems could increase the problem of antibiotic resistance. Why do we need safe thresholds on the concentration of antibiotics being released from wastewater treatment plants into the environment?
Work conducted within our group and by others has shown that very low concentrations of antibiotics, similar to those you might find in the environment can increase antibiotic resistance. Some of my previous work has also shown that in some cases, these very low concentrations can increase antibiotic resistance just as much as very high concentrations, similar to those you would use in clinical treatment.
High levels of antibiotic resistance in the environment could pose health problems for humans and animals exposed to those environments. So we need safe thresholds to help reduce the rate at which antibiotic resistance may increase, to minimize the risk of exposure, and also protect the environment.
Can you describe your latest research that has led to a way of solving this problem?
One of the main barriers to regulating the release of antibiotics into the environment is that there are very few experimental data that tell us what these ‘safe’ thresholds are. This is partly because there is no standardized experimental method that can be used to generate these data, particularly in an easy, quick, and affordable way.
Our latest research describes an experimental method that can do just that.
How does your SELECT method measure which concentrations of antibiotics are likely to increase antibiotic resistance?
For the SELECT method, a community of bacteria taken from untreated sewage is grown in lots of different concentrations of antibiotics. The density of bacteria is measured over time in an automated reader. When the bacteria are rapidly growing, the antibiotic reduces their growth in a dose-dependent way – meaning the more antibiotic, the more growth is reduced.
At a single time point during this growth phase, we determine the lowest antibiotic concentration that significantly reduces the growth of the bacterial community, compared to when there is no antibiotic present.
It turns out this gives the same or very similar results to previous studies, which have found the lowest antibiotic concentration that causes significant increases in antibiotic resistance genes. So the SELECT method appears to be a reliable proxy for increases in antibiotic resistance genes.
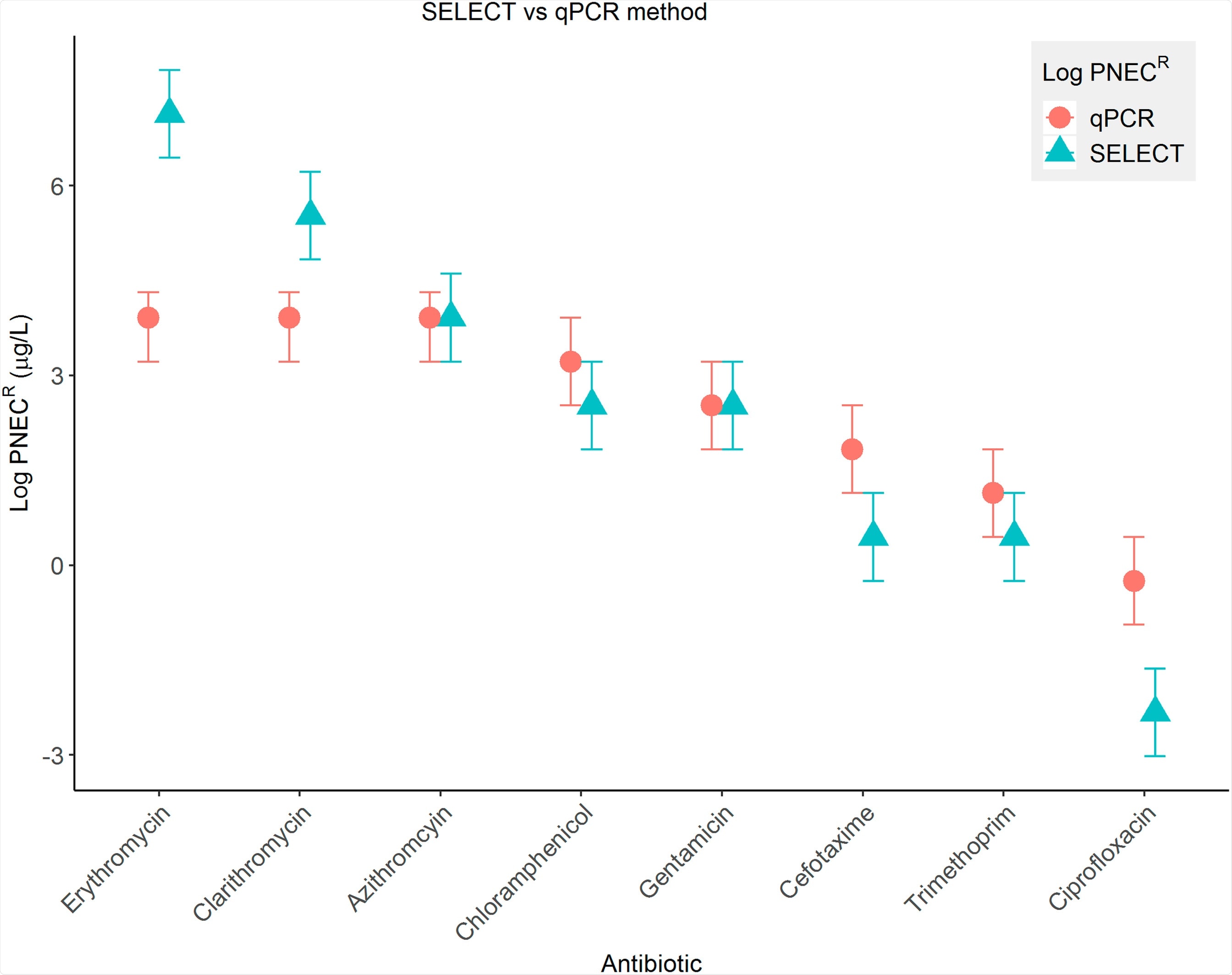
Image Credit: The ‘SELection End points in Communities of bacTeria’ (SELECT) Method: A Novel Experimental Assay to Facilitate Risk Assessment of Selection for Antimicrobial Resistance in the Environment
What advantages does the SELECT assay offer?
Compared to the methods in these previous studies, which take a week to run and two or more weeks to analyze, the SELECT method can generally generate the same or similar results in less than 15 hours. These previous methods also require specialized lab equipment and highly skilled personnel, both of which are very costly and a single experiment will cost £1000s.
The SELECT method uses minimal lab equipment and simple techniques and will cost just a few £s per run. We also found that for many antibiotics, the SELECT method is more protective than these other methods, meaning that it finds the effect of an antibiotic at lower concentrations. This is better when considering ‘safe’ thresholds, as it means we are less likely to underestimate risk and pick a threshold that might not be low enough.
The reason we see this is likely because the SELECT method takes into account all potential resistance mechanisms within that population, including mutations and entire antibiotic resistance genes; whereas the previous methods tend to only target one or two resistance genes, with every extra gene tested adding further expense.
Do you believe that by having safe measurements in place, we can help to reduce the increasing threat of antibiotic resistance?
Antibiotic resistance is an extremely complex problem that requires international efforts in so many different areas, from manufacturing and prescribing to antibiotics in the environment.
I hope that ‘safe’ thresholds for antibiotics entering the environment would help in the fight against antibiotic resistance, but no one measure will be enough on its own.
What are the next steps in your research into antibiotic resistance?
There are several! It is such a massive area, but one area we are particularly interested in is looking at mixtures of antibiotics and how they affect levels of antibiotic resistance. In the real world, you do not just have one antibiotic present at one time, but actually several, in complex mixtures alongside other chemicals that could also enrich for antibiotic resistance.
I am also interested in these other chemicals and have two Ph.D. students looking at non-antibiotic pharmaceuticals and plant protection products (like pesticides) to see if they could increase antibiotic resistance.
I also want to explore if the SELECT method can be repurposed for direct environmental surveillance of antibiotic contamination ‘hotspots’; or to re-incentivize pharmaceutical companies to move back into developing new antibiotics.
Image Credit: nokwalai/Shutterstock.com
Where can readers find more information?
About Dr. Aimee Murray
Dr. Aimee Murray was awarded a 1st Class (Hons) Degree in Biology, the Leonard Broadbent Prize and Eliahou Dangoor Scholarship from the University of Bath, 2010 – 2013.
Following the completion of her BBSRC/AstraZeneca funded Ph.D. in 2017, Aimee was awarded a NERC Industrial Innovation Fellowship to continue her research into antibiotic resistance in the environment at the University of Exeter, where she holds a proleptic lectureship due to commence summer 2021 in one of the largest research groups studying antimicrobial resistance in the environment.