In this interview, Professor Jacqueline Kimmey speaks to AZoLifeSciences about her latest research into bacterial pneumonia and the genes that may drive its spread from the lungs into the blood.
Can you give a brief overview of bacterial pneumonia and what provoked your research into this disease?
Bacterial pneumonia is a generic term for an infection of the lungs caused by a bacterium (as opposed to viral pneumonia, which is caused by a virus). Bacterial pneumonia can be caused by a variety of different species, but the most common cause is Streptococcus pneumoniae.
S. pneumoniae is a very common colonizer of the upper respiratory tract, typically without any symptoms. However, S. pneumoniae also causes invasive disease, such as pneumonia and sepsis – though it is not clear what causes this switch. I find this fascinating, that the same organism can cause life-threatening disease sometimes, and at other times, go completely unnoticed.
Image Credit: Kateryna Kon/Shutterstock.com
Why is bacterial pneumonia more common and more deadly after a viral infection? How does bacterial pneumonia interact with the flu virus?
That is a great question, but unfortunately, does not have a simple answer. Many studies have investigated this and discovered many contributing factors – including changes in the cellular environment in your lungs – which can alter the host’s ability to fight off bacterial infections as well as the nutrients available for S. pneumoniae to replicate.
S. pneumoniae can physically interact with the flu virus, which may influence its ability to spread, i.e. from the nasopharynx to the lungs or to a new host. In our new study, we found that the flu infection leads to increased susceptibility to S. pneumoniae infection – resulting in increased bacterial dissemination from the lungs to the blood.
In your research, you developed a new gene analysis method to identify the genes that drive bacterial pneumonia. Can you describe how you carried out this research?
This work utilized a CRISPR-interference to test the contribution of bacterial genes to pathogenesis. To do this, we engineered bacterial mutants that express a mutated Cas9 enzyme and guide RNA, which together act to inhibit the expression of a gene.
Importantly, the expression of Cas9 is under the control of a tetracycline-inducible promoter - which allows us to “turn off” genes by adding tetracycline, a common antibiotic. While each bacterial cell only turns off one gene (or gene cluster), we used a mixed bacterial library that targets the entire genome. So, the library is made up of bacterial cells that lack gene 1 or gene 2 or gene 3 … gene 1499, etc.
We mixed these together and infected mice – and then saw what genes are required for the bacteria to infect the lungs and the blood of mice. For example, if we find lots of bacteria in the blood that are missing gene 1, it tells us gene 1 is not required in the blood.
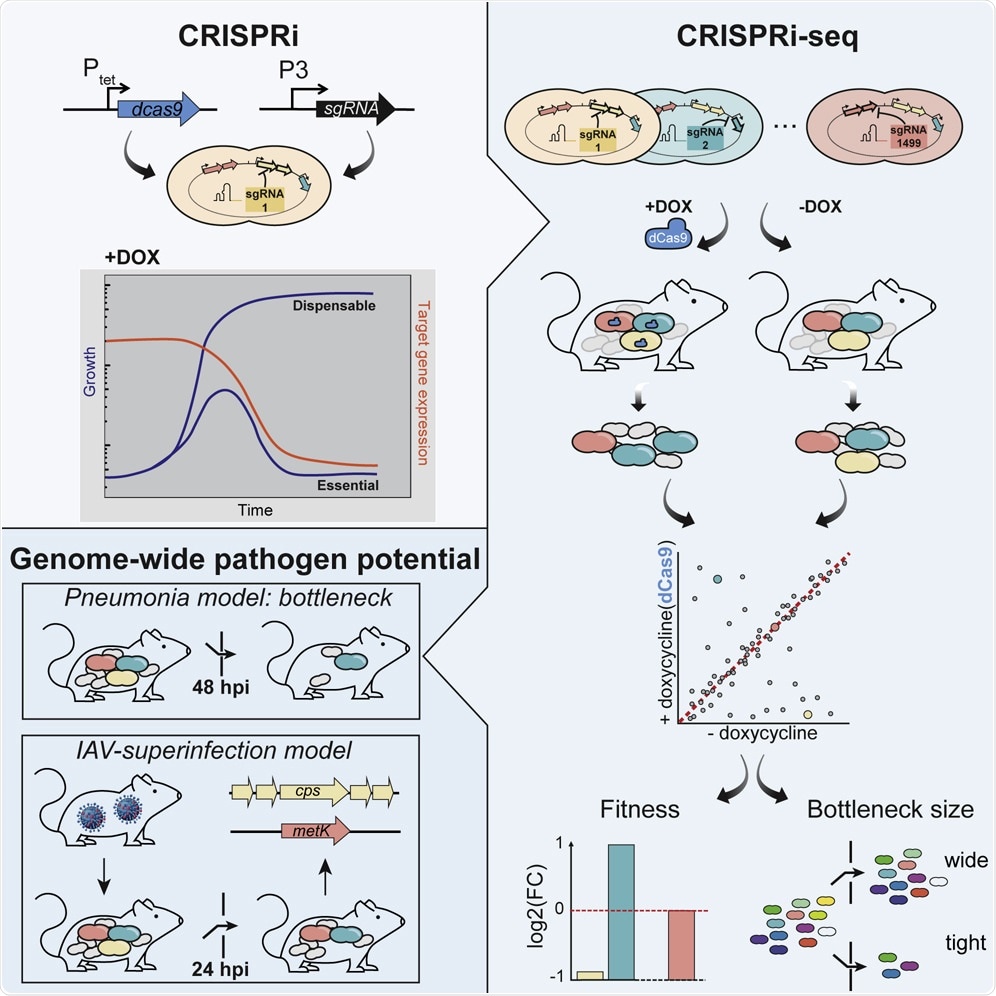
Image Credit: Exploration of Bacterial Bottlenecks and Streptococcus pneumoniae Pathogenesis by CRISPRi-Seq
What did you discover?
The most striking result was that there is a huge bottleneck that limits the number of bacteria that can get from the lungs to the blood. Even when we gave mice quite a high load of bacteria, there was a huge bottleneck, and very few bacteria, sometimes less than 25, initially make it into the blood.
Once inside the blood, these bacteria are able to replicate to very high levels. We were also surprised by the lack of consistency in terms of which strains made it into the blood, suggesting some of the disease progression is stochastic.
Interestingly, when we first infected mice with influenza A virus and then S. pneumoniae, this bottleneck was not seen, which may explain why secondary bacterial pneumonia is often more severe clinically. In addition, we were surprised to see that the main toxin, pneumolysin, was not required for the development of infection.
What role did CRISPR play in your latest research?
CRISPR uses an enzyme called Cas9 to cut DNA, commonly used for genetic engineering. In this system, we use CRISPRi which uses a dead Cas9 unable to cut DNA and instead blocks the production of the gene of interest. This is important because some genes may be required for growth in the lab (in vitro) and therefore you cannot delete the gene without killing the bacteria.
However, some of those genes may not be required during infection in a living host (in vivo), so by using our inducible system, we can turn off genes only during infection. In fact, this system allowed us to identify genes, such as metK, which are required in laboratory conditions, but not in mice.
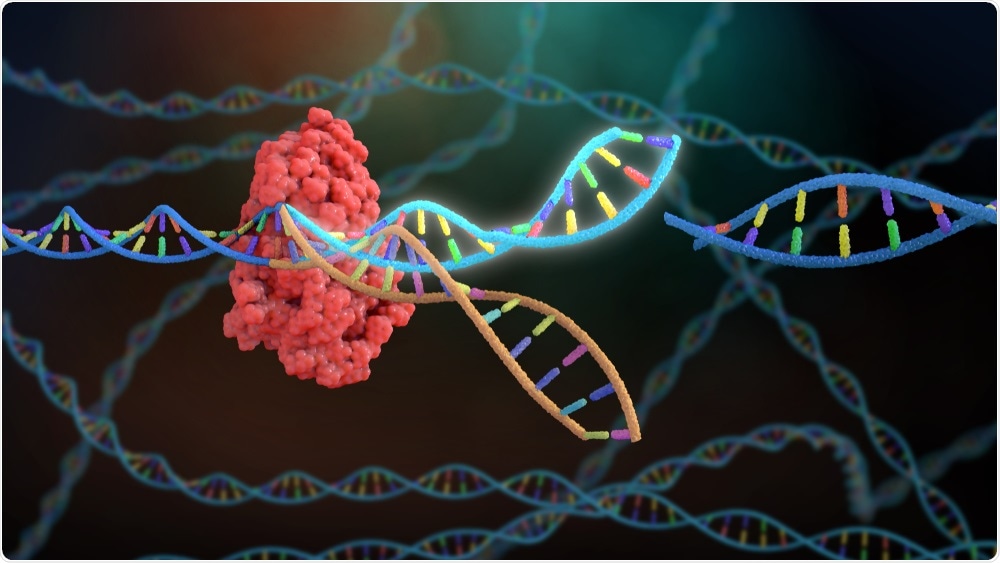
Image Credit: Nathan Devery/Shutterstock.com
Why is it important to be able to accurately identify and analyze the genes involved in infection?
It is important to know what genes contribute to infection so that we can develop therapeutics to treat disease. Continuing the above example, if you only developed antibiotics in vitro, you may find that an antibiotic that blocks metK is extremely effective in the lab, leading to rapid death of the bacteria.
However, as metK is not required in vivo, the antibiotic would not work for treatment, since metK is not essential in vivo.
What is meant by the term ‘bottleneck’ in virology and how does this bottleneck limit the volume of bacteria that enters the blood?
A bottleneck refers to a point of limitation – in infectious disease, this generally is used to refer to a stage of infection that restricts the number of infectious organisms (bacteria or viruses) that make it to the next stage.
In this study, we found a bottleneck between the lung infection and bloodstream infection – i.e. we added 1499 different bacterial strains into the lung, but less than 25 on average made it into the blood.
In your research, you noticed a lot of variation in the size of the bottleneck. Why was this exciting to see in such a highly controlled system?
Disease outcome within a population is highly variable – most people who are exposed to S. pneumoniae do not get sick, though, in others, S. pneumoniae causes life-threatening invasive disease. In general, we tend to assume this is due to population-level differences – i.e. some people may be exposed to a higher dose, may have underlying conditions, have different levels of resistance.
However, our highly controlled laboratory studies do not have any of those variables – mice are identical in terms of genetics, environment, handling, and infectious dose.
To still see such incredible variation in disease progression suggests that dynamics within disease progression are a major, and largely overlooked contributor to disease outcome, and therefore is not something that has been widely studied or targeted therapeutically.
Image Credit: Kateryna Kon/Shutterstock.com
Do you believe that your research will help to further improve our understanding of the genes involved in bacterial pneumonia?
Yes – these studies have already enhanced our understanding of genes required during infection in different tissues as well as how this changes during flu co-infection.
What are the next steps in your research?
We are particularly interested in understanding what is responsible for the bottleneck in the lungs and why this might be disrupted by the flu. We are also exploring the bacterial genes involved in infection – is it possible that a hypervirulent bacterial strain might similarly destroy this bottleneck to gain access to the blood?
Finally, a major focus of our lab is to understand what causes the variability seen during infection, including the contribution of bacterial differences as well as host immune differences.
Where can readers find more information?
About Professor Jacqueline Kimmey
Jacqueline is currently an Assistant Professor in the Department of Microbiology and Environmental Toxicology at the University of California Santa Cruz. She started her lab (www.kimmeylab.com) in 2019.
The Kimmey Lab research focuses on understanding how differences in host immune responses impact susceptibility or resistance to bacterial infection.
Jacqueline’s career has been supported by a National Science Foundation Graduate Research Fellowship and the University of California Presidential Postdoctoral Fellowship. In 2019, she was named as a Forbes' 30 under 30 listmakers (Science).