Just three years ago, researchers from the University of California, Santa Cruz (UC Santa Cruz) had demonstrated that a long-read human genome assembly could be achieved using the same nanopore technology devised on the campus.
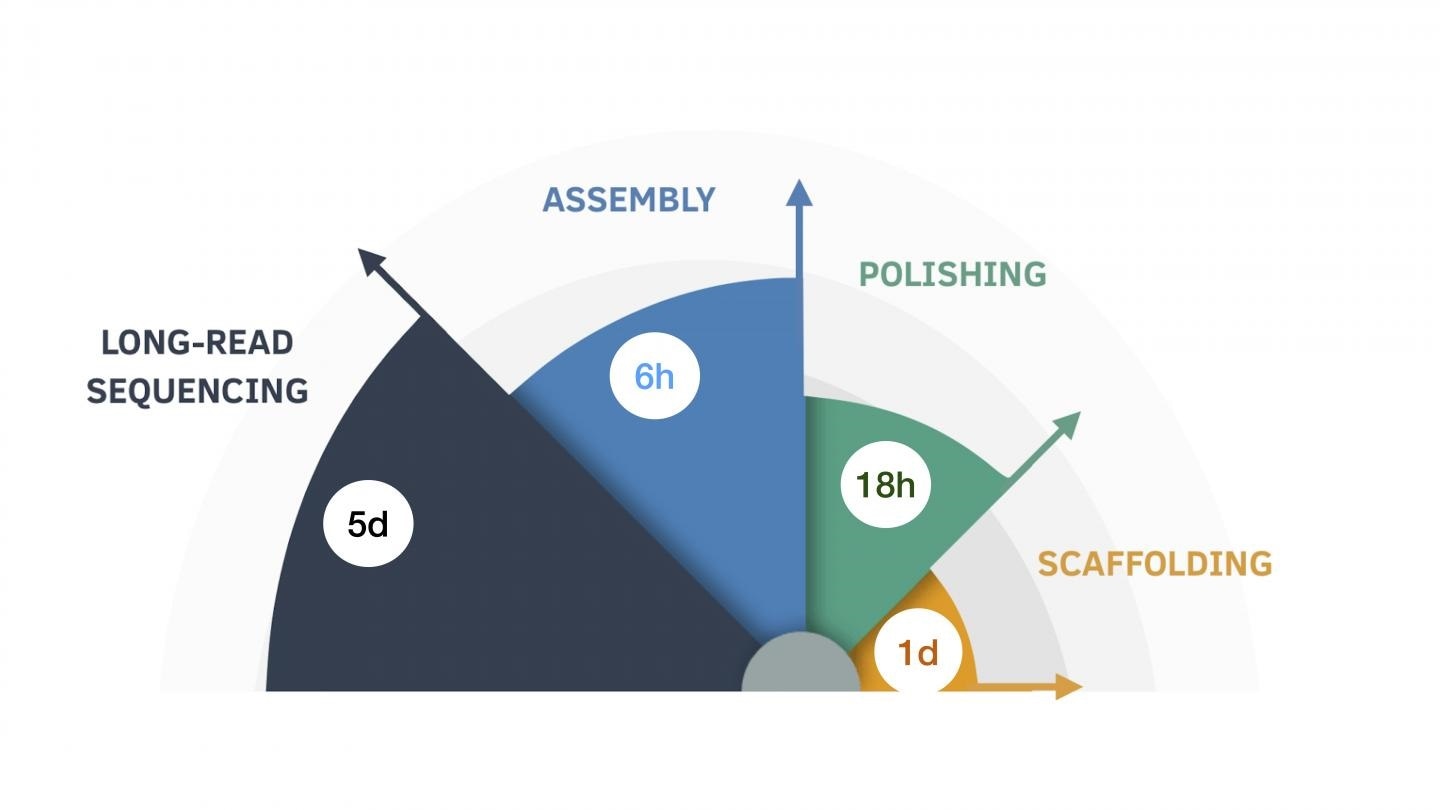
The nine-day assembly process, broken down by the length of time for each step. Image Credit: UC Santa Cruz Genomics Institute.
During that period, human genome assembly was a massive effort, involving weeks of work and as many as 150,000 hours of computing time.
Nearly a year later, the researchers used the PromethION nanopore sequencer to make an analogous effort that proved to be considerably easier, faster, and cheaper and completed the task in approximately one week.
“We sequenced eleven human genomes in nine days, which was unprecedented at the time," stated Miten Jain, a Research Scientist at UC Santa Cruz.
Now, scientists at UC Santa Cruz are working together on a new algorithm that was developed to precisely and accurately arrange complete, individual human genomes from long-read sequencing data for $70, and six hours, approximately.
According to the scientists, their assembler is expected to boost the speed of genomics research and pave the way for new opportunities. This involves allowing the pangenome research to signify the real scale of human diversity, a pursuit that is definitely more viable.
To date, genomic research has exclusively depended on the reference genome extracted from one individual chosen to denote a whole species. To mirror the real scale of human diversity, the UC Santa Cruz team has set out on a pangenomic project to sequence as many as 350 new, individual human genomes.
As part of this initiative, the scientists from UC Santa Cruz Genomics Institute have devised a nanopore long-read sequencing procedure that reliably produces approximately 60X coverage (that is, around 200 gigabases) of a human genome at unparalleled lengths (median read N50 of 42 kb), with the help of three PromethION flow cells. Moreover, around 7X coverage of the human genome is in reads, the length of which surpasses 100 kb.
This kind of technique is quite scalable, in terms of cost and also in terms of the number of human genomes that can be processed at the same time. The researchers are currently enhancing this technique for greater throughput and read lengths, which will additionally enable the goal of realizing human genomes that are phased, complete, and have reference quality.
This massive data inflow necessitated the development of extremely efficient software tools, beginning with an assembler.
Our new assembler was designed to be cheap and quick, with the goal to be on the cloud. It gives us the power to scale nanopore sequencing. Now, I'm confident that we'll be easily assembling hundreds of de novo genomes in the next couple of years.”
Benedict Paten, Assistant Professor, Department of Biomolecular Engineering, University of California, Santa Cruz
The solution was contributed by a large group of developers and scientists headed by Paolo Carnevali from the Chan Zuckerberg Initiative (CZI), and also involved many researchers at the Computational Genomics Lab at the UC Santa Cruz Genomics Institute.
“When I saw the Jain 2018 paper, I was impressed and realized that I could contribute to the computational side of this line of investigation,” stated Paolo Carnevali. “I had recently met Benedict Paten and decided I wanted to work with his team at UCSC.”
The researchers soon started to work in tandem. In just a few months, they had created and validated the unique algorithmic sauce, which they dubbed Shasta.
According to the authors, Shasta is a new, in-memory computing-driven algorithm that can presently allow scientists to complete a de novo (new and never before processed) assembly of human genomes within six hours, for an average cost of $70 for each sample.
In their article titled, “Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes,” published recently in the Nature Biotechnology journal, the researchers elucidated how the Shasta algorithm yields similar or more improved precision as its contemporaries and also has the least number of misassemblies.
However, the researchers were not content with this milestone and saw a chance to enhance the draft assembly at a cheaper turn-around time and cost.
To improve the base-level quality of the assemblies, we used a sequence polisher based on a deep neural network as the final assembly step. This brought the total cost of the assembly process to less than $200 and 37 hours—which further reduced the computational overhead of generating long-read assemblies dramatically—by a factor of five.”
Kishwar Shafin, Study Lead Author, University of California, Santa Cruz
When the scientists evaluated and verified the precision and accuracy, respectively, they observed that they had obtained an accurate assembly of 99.9% by simply using the nanopore data—the first-of-its-kind for the human genome.
Then using the HiC sequencing data, the scientists also produced chromosome-level scaffolds for those polished assemblies.
Karen Miga, a research scientist, the study’s co-author, and who is also managing the Data Production Center at UC Santa Cruz for the Human Pangenome Project, underscored the importance of the researchers’ accomplishment in enhanced accuracy.
Our aim is not only to expand the diversity of the reference genome but also to resolve the hundreds of gaps that persist across the genome. Now that we can routinely include these uncharted regions, we have a truly complete assembly of a human genome, and we can begin to explore variations of unknown consequence.”
Karen Miga, Study Co-Author and Research Scientist, University of California, Santa Cruz
Source:
Journal reference:
Shafin, K., et al. (2020) The Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nature Biotechnology. doi.org/10.1038/s41587-020-0503-6.