With the free and recently developed bioinformatic tool, called SSRgenotyper, scientists can now use simple sequence repeats (SSRs) to digitally genotype sequenced populations—a task that formerly involved time-intensive laboratory-based techniques.
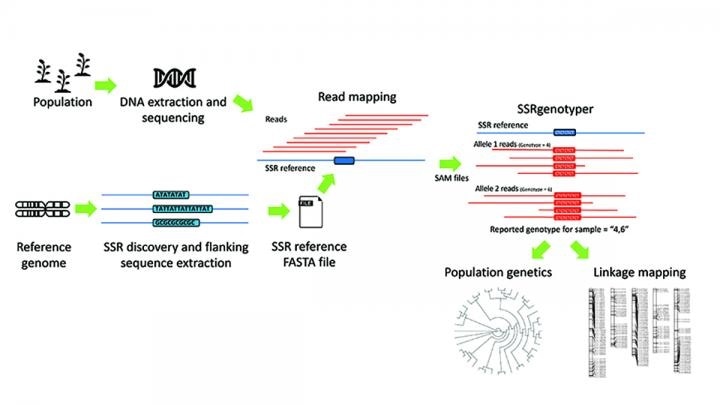
A workflow depicting the process of SSR discovery, DNA amplification, and read mapping. Once SSRs have been identified and mapped, that information can be exported as a SAM to SSRgenotyper to perform all relevant genotyping, the removal of spurious alleles, and filtering. The genotyping results can then be exported in several file times for further downstream analyses. Image Credit: Lewis, D. H., D. E. Jarvis, and P. J. Maughan. 2020. SSRgenotyper: A simple sequence repeat genotyping application for whole-genome resequencing and reduced representational sequencing projects. Applications in Plant Sciences 8(12): e11402.
The developers of the new tool have designed the program to flawlessly incorporate with other types of applications that are being used for detecting and analyzing SSRs. The researchers have reported the results in the latest issue of the Applications in Plant Sciences journal.
SSRs are essentially short chains of repeating nucleotides that have a tendency to mutate. The inconsistency of such DNA sequences makes them suitable for genetic studies to differentiate between people, and hence, are often the preferred marker for forensic testing and paternity.
When it comes to research fields, SSRS have the additional advantage of being selectively neutral, which means, they do not code for any physical traits and, hence, are not subject to several types of natural selection, rendering them an exceptional tool to investigate populations without the concealing effects of convergent evolution.
Now, recent developments in next-generation sequencing have allowed scientists to simplify the SSR identification process, particularly in groups or model organisms with an existing assembly of a reference genome.
With decreasing sequencing costs and ongoing improvements in technologies, sequencing massive genomic portions for the purpose of SSR studies, even in the case of non-model organisms, are turning out to be more viable and extensive in the scientific literature.
But the process of genotyping—that is, establishing which persons have which alleles—still depends mainly on observing amplified DNA on an electrophoresis gel, which happens to be an intensive and potentially dangerous process, as fragments of DNA are usually stained with carcinogenic chemicals.
In this process, another problem is that alleles are quantified depending on the size of the ensuing bands, which is an approximation of the total number of nucleotides present in the amplified DNA fragments.
Since there is likely to be a subtle difference in the flanking regions surrounding the target SSRs, and because there is a lack of standardized technique to establish the size of an allele through these techniques, genotyping outcomes from just a single experiment cannot be simply transferred or compared to the results of another experiment.
Such laboratory-based efforts have become absolute because of the development of SSRgenotyper. When combined with other bioinformatics programs that identify SSRs in reference DNA as well as programs that align sequence information from target populations with the equivalent SSR reference file, the SSRgenotyper can rapidly genotype all SSRs for all separately sequenced specimens.
SSRgenotyper goes the next step by genotyping SSRs within sequenced populations -- strictly from sequencing data (no PCR or electrophoresis). The output from SSRgenotyper are files ready for population genetic analysis or linkage map formation.”
Jeff Maughan, Study Senior Author and Professor of Plant and Wildlife Sciences, Brigham Young University
The program not only decreases the amount of work and the time needed to genotype populations, but it also resolves the transferability issue integral in the estimation of electrophoresis by instantly counting the overall number of base pairs in a specified sequence repeat.
Since the SSRs are genotyped based on the number of repeated motifs at the SSR locus and not on the PCR product size, the allele calls are standardized and transferable from project to project or from lab to lab.”
Jeff Maughan, Study Senior Author and Professor of Plant and Wildlife Sciences, Brigham Young University
Coded in Python 3, the program needs just three positional arguments to run and offers the choice to specify many conditional arguments (like percentage thresholds for the size of the flanking regions, heterozygosity, and the elimination of false alleles). Moreover, the program can be conducted on a standard desktop computer.
As soon the SSRgenotyper is over, it creates multiple types of files, such as statistical files and basic summary, and also a .map, a .pop, and an alignment file that is formatted for use in more programs to enable downstream analyses.
As a proof of concept and to test the ability of the SSRgenotyper in precisely determining a person’s genotype, Maughan and his collaborators ran the program on freely available sequences of the oat Avena atlantica and quinoa (Chenopodium quinoa) species. The accuracy rate, which was thus obtained, was 97% or more, which increased on adding more sequence reads.
With the efficiency and the ongoing development of next-generation sequencing techniques, tools such as SSRgenotyper are set to decrease the amount of laboratory work needed in genetic studies.
Sequencing is already the method of choice in most genetic research projects. As costs continue to drop and new bioinformatic tools are developed, it is highly likely that future population genetics studies will be based solely on next-generation sequencing—completely avoiding the cumbersome tasks of PCR and electrophoresis.”
Jeff Maughan, Study Senior Author and Professor of Plant and Wildlife Sciences, Brigham Young University
Source:
Journal reference:
Lewis, D. H., et al. (2020) SSRgenotyper: A simple sequence repeat genotyping application for whole‐genome resequencing and reduced representational sequencing projects. Applications in Plant Sciences. doi.org/10.1002/aps3.11402.