Researchers at Tokyo Metropolitan University have discovered an exclusive mechanism through which two transcription factors stabilize each other’s binding to DNA in fission yeast.
Transcription factors Atf1 and Rst2 help stabilize each other and bind to an opened segment of the chromatin structure. Their proximity, a mere 45 base pairs apart, is key to this effect, as demonstrated by the team’s experiments. Image Credit: Tokyo Metropolitan University.
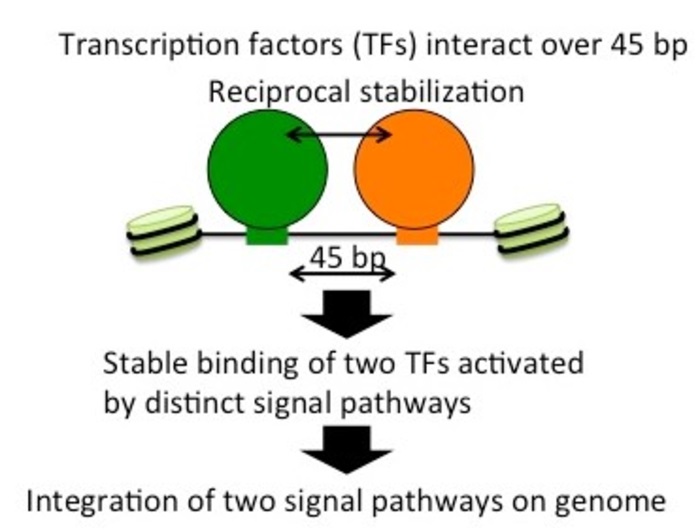
They identified that Atf1 and Rst2 enable each other to stably bind upon coming sufficiently close together. They both enable transcription of a gene that handles glucose poor environments but fall in completely independent activation pathways. Such a novel understanding can help researchers in their fight against cancer.
The familiar visualization of the DNA helix resembles a long winding molecular thread including all the information needed to develop and sustain life. However, a not-so-known fact is that it is neatly packaged and stored within cells: DNA is wound around protein structures called histones, thereby creating an elegant, tightly packed structure called the chromatin.
For molecular processes to use that information, the chromatin “opens,” thus rendering the DNA available for binding by transcription factors, which are proteins helping in the translation of the DNA sequence made of base pairs (or “letters”) into messenger RNA (mRNA). Then, the mRNA is finally read by a ribosome to synthesize proteins based on the original blueprint.
The way transcription factors (TF) bind to the chromatin is a crucial focus of biomedical research. Several types of cancers, for instance, originate when this process goes wrong. A research team headed by Prof. Kouji Hirota of Tokyo Metropolitan University has been investigating this process by analyzing a simpler organism, the fission yeast. Their focus is on its response to changes in its environment.
The researchers have now successfully identified the unique mechanism of how transcription works in yeast cells that respond to a lack of glucose in their environment.
In starving yeast cells, transcription of the fbp1 gene is known to be massively stimulated by two TFs—Atf1 and Rst2. The researchers explored this process in-depth and identified not only that the activation of both was critical to the function of fbp1 but also that they actually helped stabilize each other.
They could explicitly demonstrate that this was mainly due to how close these sites were, often only 45 base pairs apart. The introduction of additional lengths of DNA between the sites suddenly affected the ability of the TFs to help each other, and the chromatin closed. This left both factors unbound. The relative orientation of the TFs along the twisting grooves of the helix was also found to be crucial.
Most significantly, this effect was demonstrated to be sufficiently strong to counteract the effects of Tup11 and Tup12—co-repressors which play a role in destabilizing the random binding of independent TFs to the chromatin. All this indicates that this reciprocal relationship not just helps the TFs bind successfully but also prevents both from attaching by themselves.
The strange thing is that these TFs are stimulated by totally independent chemical pathways. The process identified by the researchers thus combines these routes together into a signal “hub.”
This finding is just a single piece in a complex biochemical puzzle but still helps emphasize an unappreciated mechanism used by different TFs to interact and effectively combine pathways together. The researchers believe this new understanding can help in their fight against cancer and other associated illnesses.
Source:
Journal reference:
Koda, W., et al. (2021) Reciprocal stabilization of transcription factor binding integrates two signaling pathways to regulate fission yeast fbp1 transcription. doi.org/10.1093/nar/gkab758.