Proteins attach to one another through a complex mixture of chemical interactions, but what if the process is simpler—like binding due to their shapes? To nail the answer to this question, scientists employed Summit—the nation’s fastest supercomputer—to model lock-and-key interactions.
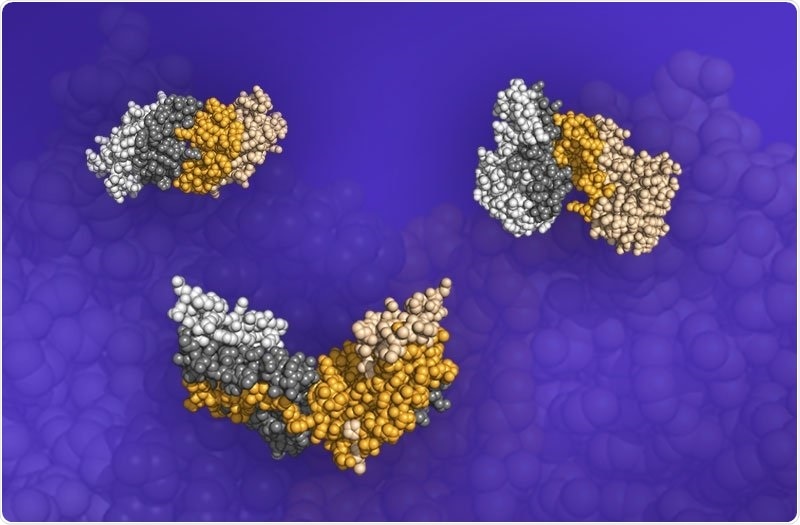
Three protein pair structures. The darkened portions of the structures are the interfaces where the proteins meet and bind. Image Credit: Image courtesy of Fengyi Gao, University of Michigan and Oak Ridge National Laboratory.
In these interactions, molecules present in the proteins attaching together should chemically “fit” accurately. The researchers analyzed 46 protein pairs that are known to attach to one another.
As a further step, the researchers used Summit to model the assembly of protein pairs. Of the 46 pairs examined, six assembled depending on their complementary shapes more than 50% of the time.
The impact
The results can induce numerous applications in biological research. For instance, this approach can offer researchers information on the usage of proteins as building blocks to create novel biological materials or screen drugs for disease treatments.
The group intends to analyze more proteins that can also bind depending on shape—or even shape higher-order assemblies. Eventually, the researchers want to find the restriction for the protein shapes evolving to form hierarchical protein structures.
Summary
Successful binding of proteins to one another requires one of them acting as a ligand—a molecule binding to a target protein—and one of them acting as a receptor—a molecule receiving the ligand. This mechanism involves complex chemical interactions where the molecules share bonds and alter their configurations after binding.
Scientists wanted to analyze if they can foretell this binding depending on shape alone, overlooking protein interactions. The researchers analyzed 46 pairs from a database of above 6,000 protein pairs. The analyzed pairs are known to attach to one another and they simulated their assembly on Summit, the Oak Ridge Leadership Computing Facility’s 200-petaflop supercomputer.
The researchers captured six protein pairs that attached depending based on shape alone, with one of the pairs binding more than 94% of the time. The researchers intend to analyze more proteins that too can bind based on shape—or even form higher-order structures.
The researchers also intend to comprehend the limitation of how protein shapes can evolve to form hierarchical protein structures in the future. The researchers anticipate that they can ultimately foretell the attachment of protein-protein interfaces in protein crystallization structures or protein clusters.
Source:
Journal reference:
Gao, F., et al. (2021) The role of complementary shape in protein dimerization. Soft Matter. doi.org/10.1039/D1SM00468A.