In this interview, we speak to Dr. Chao Ma, one of the speakers at SLAS 2022, about his groundbreaking 'leukemia-on-a-chip' technology and its future within therapy resistance.
Please could you introduce yourself and tell us what inspired your career in the life sciences?
Thank you for inviting me to discuss our recent progress on “Leukemia-on-a-Chip” for elucidating chemoresistance and drug testing. My name’s Chao Ma. Currently, I am completing postdoc training at New York University Tandon School of Engineering. Since 2017 when I first came to the US and started working at Tandon, I have been leading a group of junior students on in vitro modeling of tumor micro-environments to understand therapy resistance and screen for optimal therapy, for example, chemoresistance and CAR-T immunotherapy.
When I was a high school student, I was fascinated by the mystery of biology as well as the beauty of engineering, so I applied to an innovative and honor program from Northwest A&F University (China), which allows students to pursue their undergraduate and graduate study in a successive period of 8 years. During that period, honestly, studying was hard and boring. But at one critical moment when I was in the class of animal histology and embryology, I was shocked by the beauty of liver tissue composed of numerous hexagon-like tissue units and liver lobule which, when working together, can help the human body metabolize various biochemical and of course drugs.
I also realized that engineering platforms, i.e., organ-on-a-chip, I learned from another class could help to build this beautiful and delicate liver unit in vitro helping to screen out drugs that are toxic and not disease-treating. Since then, I devoted myself to micro-engineering in vitro liver tissue for analyzing metabolism-related drug toxicity, later pursuing an academic career in vitro models.
Image Credit: Kateryna Kon/Shutterstock.com
You are giving a talk at SLAS 2022 in Boston titled ‘Leukemia-on-a-Chip for modeling and decoding the heterogeneous mechanisms underlying therapy resistance’. Can you give us a brief overview of what you will be covering in this talk and what visitors can expect?
Yes, of course. First of all, I will introduce how this in vitro organotypic “Leukemia-on-a-Chip” model is designed and engineered to emulate the in vivo B-ALL BM pathology. Following this, I will show how the leukemic perivascular, endosteal, and hematopoietic niche-derived factors maintain B-ALL survival and quiescence within a spatial and genetic heterogeneity pattern. Finally, I will demonstrate how our model can be used in the preclinical setting to test niche-co-targeting regimens, which may translate to patient-specific therapy screening and response prediction.
Those who are interested in tissue engineering, disease modeling, and drug discovery are expected. Also, those who are interested in microfluidics, automatic system engineering, and high-throughput drug testing are welcomed.
B-cell acute lymphoblastic leukemia (B-ALL) can sometimes become resistant to therapy. Can you give an overview of this type of leukemia and how its resistance occurs?
B-cell acute lymphoblastic leukemia (B-ALL) is the most common cancer among children and is characterized by the overproduction of immature and dysfunctional B cell blasts within bone marrow (BM). Despite the substantial progress achieved over the past decade with multidrug chemotherapy regimens, allogeneic hematopoietic stem cell (HSC) transplantation, and, most recently, CD19-targeted CAR (chimeric antigen receptor) T cell immunotherapy, relapse is common after initial treatment and the leading cause of death for pediatric patients with B-ALL.
Patients with refractory and relapsed B-ALL have a poor prognosis with a 5-year survival rate of about 10%, largely because of acquired resistance by the heterogeneity in the BM microenvironment and tumor genetics. Currently, there is limited prognostic information between these heterogeneity and therapeutic responses, such as patients with ETV6-RUNX1+ B-ALL often have favorable outcomes, while patients with Philadelphia chromosome-positive (Ph+) B-ALL are expected to have an unfavorable prognosis.
A clearer understanding of the microenvironmental evolution during leukemia pathogenesis and the heterogeneity of acquired chemoresistance mechanisms from distinct B-ALL subtypes may provide previously unidentified therapeutic targets for optimized therapy for patients with refractory and relapsed B-ALL.
Despite its resistance, studying its interactions is currently limited by in vivo methods. Why is this and how does your new ‘leukemia-on-a-chip’ device overcome this challenge?
At present, all preclinical murine models, with the aid of intravital microscopy, have provided the foundation of the in vivo leukemic BM ecology. Results emanating from these approaches may be difficult to reproduce as demonstrated, for instance, in previous T-ALL studies highlighting differences in leukemic blast localization in vivo, there was difficult translating it to molecularly distinct B-ALL subtypes. Moreover, scanning cell compositions in leukemic BM niches provide an overall demographic of BM cell populations, but it does not map the real-time and dynamic evolution of the tumor-niche cross-talk.
The conventional two-dimensional (2D) or 3D cell co-culture systems are simple and convenient platforms for biological studies but they cannot recapitulate the key architectures and characteristics of the in vivo B-ALL BM niche such as the central sinus, medullar cavity, and endosteal space as well as the hematopoietic environment.
Our microfluidics-based microphysiological system recapitulates key features of in vivo BM tissue architecture and is able to monitor dynamic B-ALL and leukemic BM niche interactions in real-time with live-cell imaging, which is difficult for traditional animal studies. Moreover, our biomimetic system is able to detangle interactions between B-ALL blasts and its leukemic BM niche within in vitro central sinus, medullary cavity, and endosteum anatomical regions as well as the hematopoietic environment.
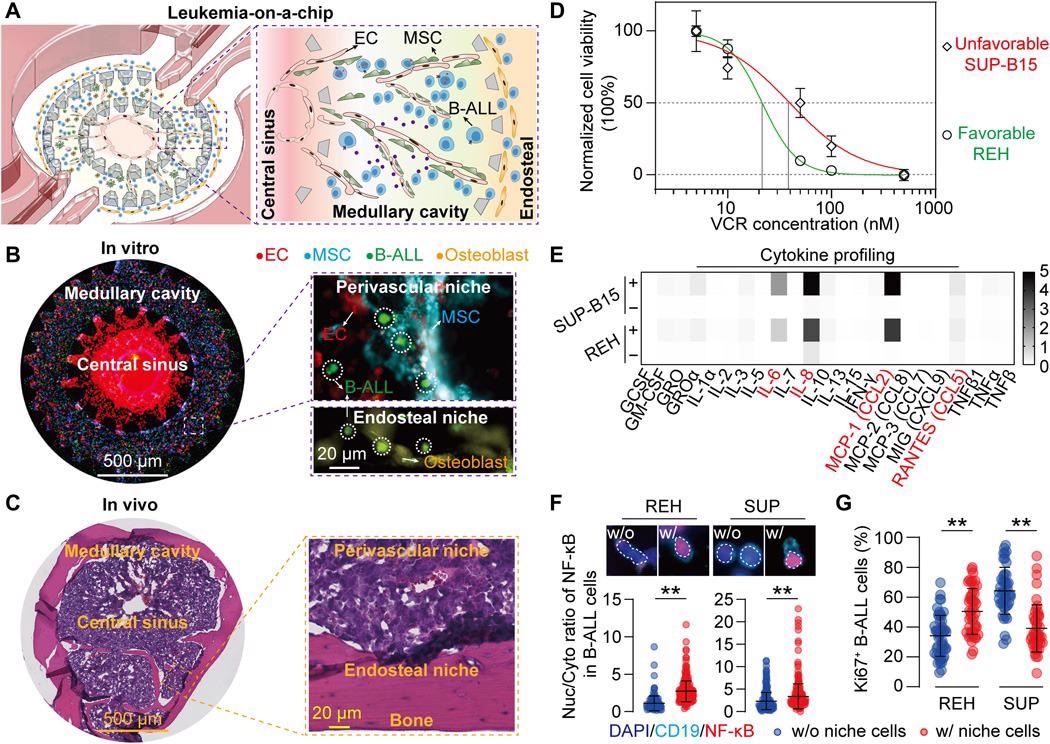
Image Credit: Leukemia-on-a-chip: Dissecting the chemoresistance mechanisms in B cell acute lymphoblastic leukemia bone marrow niche
Can you describe how you designed and made your latest ‘leukemia-on-chip-device’? How does it work?
Our design of ‘Leukemia-on-a-Chip’ device is biologically inspired. As we know, the bone marrow is vascularized with an arterial supply ramifying from the center into the vascular network (medullary cavity), merging with sinusoids at the peripheral region (endosteal region) then back into a central sinus.
To mimic this concentric and layered structure of bone marrow, we design a multi-compartmentalized microfluidic device that is divided into a central venous sinus (center region), a medullary cavity (middle ring), and an endosteal region (outer ring). These three functional regions were partitioned by regularly spaced trapezoid micropillars that confine cell-embedded hydrogels, by balancing surface tension and capillary forces, and were also connected with four medium reservoirs for long-term medium supply.
The sequenced loading of different cell types in a biomimetic hydrogel solution into different compartments of the microfluidic device will thus reproduce the tissue structure of bone marrow. Self-assembly of B-ALL blasts and its niche cells during the culture period will further remodel the engineered system into a biomimetic leukemic bone marrow niche.
What did you discover when monitoring the behavior of B-ALL blasts? How will your device help to design future therapies for leukemia?
We first found that B-ALL blasts attracted endothelial cells during the 3-day culture. After we quantified the intercellular distance between B-ALL and niche cells during co-culture, we found that B-ALL and niche cells dynamically co-localized. We further observed an enhanced clustering of B-ALL cells around niche cells compared to the B-ALL monoculture condition, indicating that niche cells may provide additional adhesive sites to facilitate B-ALL clustering.
Time-lapse migration analysis of B-ALL cells revealed that leukemic cells either cultured in the presence or absence of niche cells were comparably motile at the culture onset (day 0), whereas after a two-day culture, leukemia blasts co-cultured with niche cells were less motile, as compared to those cultured in the absence of niche cells, suggesting the temporally evolved features in chemotactic and adhesive signaling among B-ALL blasts and niche cells.
B-ALL is one of the most common types of leukemia, the most common cancer in children, and has distressing consequences including increased susceptibility to infections and the need for chemotherapy and treatment. How could your research be applied and transform the lives of these children?
Yes, correct. Most children of cancer patients suffer from B-ALL. Our ‘Leukemia-on-a-Chip’, when infused with patient-derived cells, will be able to recapitulate patient-specific characteristics, which will allow us to evaluate a patients’ response to a given therapy prior to the clinical treatment. This evaluation will help physicians to make better decisions on which regiment and prescription should be given to patients.
Herein, I would like to talk about another possibility that our ‘Leukemia-on-a-Chip’ can deliver and is currently under development in our lab at NYU.
More recently, CAR T cells have emerged as a promising Food and Drug Administration–approved immunotherapy for relapsed and refractory B-ALL. However, patient responses are largely unpredictable. A detailed understanding of the leukemic BM immune niche is also indispensable for improving CAR T cell therapy.
By increasing the biological complexity of our ‘Leukemia-on-a-Chip’ platform with the addition of patient-derived cells, such as immune cells, to answer how the BM immune niche-derived regulatory signals influence leukemia progression and clinically relevant immune resistance to CAR T-cell immunotherapy.

Image Credit: Photographee.eu/Shutterstock.com
Do you believe that your technology could be repurposed and used to study other hematological malignancies also? What benefits would this have within drug discovery?
Yes, our current Leukemia-on-a-Chip platform is a tissue-engineered model of bone marrow that is inhabited by B-cell acute lymphatic leukemia. This bioengineered platform can be immediately applied to many other hematological malignancies with little to no modification.
For instance, AML, diffuse large B cell lymphoma, and multiple myeloma, as well as to study HSCs in a healthy BM niche, presenting an enabling tool for drug development and treatment testing under more physiologically relevant contexts.
As technology continues to evolve and emerge within science, what are you most looking forward to?
I would like to see a vista that as technology continues to evolve and emerge within science, the Organ-on-a-Chip technology can be organically incorporated into the drug development pipeline from early drug discovery to preclinical screening, testing, and translation of new drugs, which bridges the gap between animal studies and clinical trials involving human subjects.
What does the future of microfluidic devices look like to you? Do you see these devices becoming more prominent within the life sciences?
In my opinion, the future development of microfluidic devices will continuously integrate novel engineering tools (e.g., automation handling, 3D printing, and in situ multi-sensors) and biological concepts (e.g., patient-specific induced pluripotent stem cells and organoids) into a personalized Organ-on-a-Chip platform that will unprecedentedly promote its broad applications in biomedical, translational, and clinical scenarios.
I have seen a trend in biological studies that a lot of biologists have adapted organ-on-a-chip platforms to validate their results before and after running animal tests.
Where can readers find more information?
Our publication of “Leukemia-on-a-Chip” in Science Advances, https://www.science.org/doi/10.1126/sciadv.aba5536
Our opinion on the role of organ-on-a-chip platform for drug development, https://www.sciencedirect.com/science/article/pii/S0165614720302649
About Dr. Chao Ma
Dr. Ma is currently a Cancer Research Institute Irvington Postdoctoral Fellow at New York University. His current researches focus mostly on engineering Leukaemia-on-a-Chip platform to understand resistance to chemotherapy and recently CAR T-cell immunotherapy. Dr. Ma received his B.S. in Biotechnology in 2013 and Ph.D. in Animal Biotechnology (Cellular Engineering) in 2017 both from Northwest A&F University.
During that time, he developed micro-engineering strategies to construct liver tissue in vitro for analyzing metabolism-related drug toxicity and bottom-up liver tissue engineering. Dr. Ma has been awarded nationally and internationally, such as National Scholarship for Encouragement Award from College of Innovation and Experiment, NWAFU (2021), National Scholarship for Graduate Students Award from the Ministry of Education, China (2016), Outstanding Graduates Award from College of Veterinary Medicine, NWAFU (2017), Irvington Postdoctoral Fellowship Award from Cancer Research Institute (2021), DMM Conference Travel Grant Award from the Company of Biologists (2021), Post-Doctoral Researcher Travel Award from BMES-CMBE (2022), as well as Tony B. Academic Award from SLAS (2022).
Dr. Ma has emerged as an expert in the microfluidic Organ-on-a-Chip field with invitations to present his findings at local, national, and international conferences (WPC, BMES, ASME, CMBE, and SLAS). Dr. Ma is also a co-inventor on two patents involving engineering methods for tissue and tumor modeling for advanced therapy and is the author of numerous peer-reviewed publications.
The overarching research goals for Dr. Ma are to develop and leverage a multidisciplinary approach that combines engineering with biology and medicine for tissue engineering, disease modeling, and therapy screening, which aims to positively impact human healthcare and wellness.