CRISPR has marked the beginning of a new era in genomic medicine. The popular CRISPR-Cas9 has generated plenty of potent methods for curing genetic diseases. However, there is a last-mile problem: these tools must be given to every cell in the patient, and most Cas9s are too large to fit into common genome therapy vectors like the adenovirus-associated virus (AAV).
 A comparison of a Cas9 molecule, left, and an IscB molecule, right. Image Credit: Cornell University.
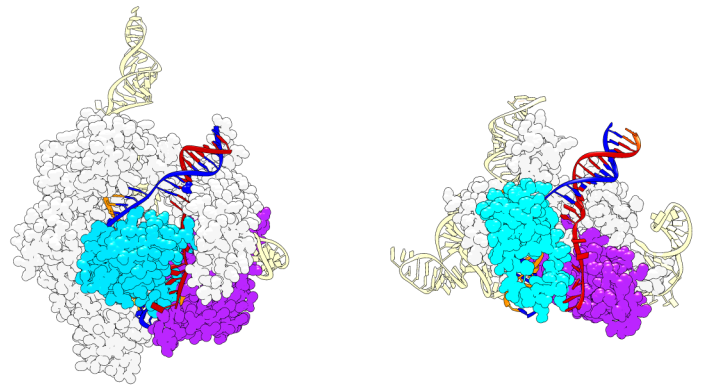
A comparison of a Cas9 molecule, left, and an IscB molecule, right. Image Credit: Cornell University.
Cornell scientists have published new research that explains how nature solves this problem: scientists specify with atomic precision how a transposon-derived mechanism changes DNA in an RNA-guided manner. Inside bacteria, transposons are mobile genetic elements. IscB, which is less than half the size of Cas9 but similarly capable of DNA editing, is encoded by a transposon lineage. Cas9 would be replaced with IscB, which would solve the size issue completely.
The study was published in Science on May 26th, 2022.
The IscB-ωRNA molecule from a transposon system was seen in high resolution using cryo-electron microscopy (Cryo-EM). Researchers were able to take pictures of the system in various conformational states. By deleting non-essential elements from IscB, they were able to build smaller IscB variations.
Next-generation fancy applications require the gene editor to be fused with other enzymes and activities and most Cas9s are already too big for viral delivery. We are facing a traffic jam at the delivery end. If Cas9s can be packaged into viral vectors that have been used for decades in the gene therapy field, like AAV, then we can be confident they can be delivered and we can focus research exclusively on the efficacy of the editing tool itself.”
Ailong Ke, Study Corresponding Author and Professor, Molecular Biology and Genetics, College of Arts and Sciences, Cornell University
CRISPR-Cas9 systems detect a DNA sequence using an RNA guide. When a match is detected, the Cas9 protein snips the target DNA at the correct spot, allowing for genetic disease surgery at the DNA level.
The Cornell team’s cryo-EM findings suggest that the IscB-ωRNA system functions in a similar way, with the Cas9 protein’s decreased size achieved by replacing sections of it with a structured RNA (ωRNA), joined to the guide RNA. The IscB protein is shortened to the core chemical reaction sites that snip the target DNA by replacing protein components of the bigger Cas9 with RNA.
It’s about understanding the molecules’ structure and how they perform the chemical reactions. Studying these transposons gives us a new starting point to generate more powerful and accessible gene editing tools.”
Gabriel Schuler, Study First Author and Doctoral Student, Graduate Field of Microbiology, Cornell University
Transposons, or mobile genetic elements, are thought to be the evolutionary precursors of CRISPR systems. Nobel Laureate Barbara McClintock ‘23, M.A. ‘25, PhD. ‘27 found them.
Ke added, “Transposons are specialized genetic hitchhikers, integrating into and splicing out of our genomes all the time. The systems inside bacteria in particular are being selected constantly—nature has basically tossed the dice billions of times and come up with really powerful DNA surgical tools, CRISPR included. And now, by defining these enzymes in high resolution, we can tap into their powers.”
The researchers anticipate they will be able to decrease IscB even further when compared to CRISPR Cas9. They have already reduced 55 amino acids from IscB without harming its function, and scientists intend to make future versions of the genome editor considerably smaller and more helpful.
Another goal of the research was to better understand the function of the companion guide RNA.
There’s still a lot of mystery – like why do transposons use an RNA-guided system? What other roles this RNA may be playing?”
Chunyi Hu, Study Co-First Author and Postdoctoral Researcher, Department of Molecular Biology and Genetics, Cornell University
While the IscB-ωRNA is incredibly active in test tubes, it is not as effective at modifying DNA in human cells, which remains a hurdle for researchers. The next phase in their study will be to use the molecular structure to investigate the causes of low activity in human cells that they have discovered. “We have some ideas, a lot of them actually, that we are eager to test in the near future,” Schuler concluded.
Source:
Journal reference:
Schuler, G., et al. (2022) Structural basis for RNA-guided DNA cleavage by IscB-ωRNA and mechanistic comparison with Cas9. Science. doi.org/10.1126/science.abq7220.