One of the most crucial techniques for examining biological activity in cells is single-cell RNA sequencing (scRNA-seq), but it is constrained by the possibility that the data it produces may be inaccurate. A research team from Japan has now created a brand-new technique called terminator-assisted solid-phase complementary DNA amplification and sequencing (TAS-Seq), which gets over these restrictions and offers more precise data than the current scRNA-seq systems.
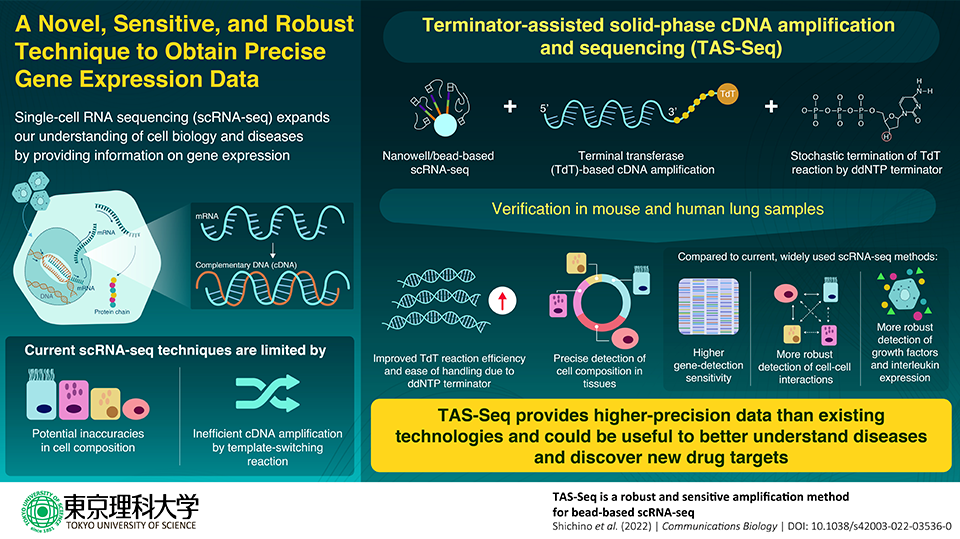 The new technique employs a nanowell/magnetic bead-based single-cell RNA sequencing platform, terminal transferase enzyme, and dideoxynucleotide phosphate terminators to generate high-precision information on gene expression, cell composition, and cell–cell interactions. Image Credit: Tokyo University of Science.
The new technique employs a nanowell/magnetic bead-based single-cell RNA sequencing platform, terminal transferase enzyme, and dideoxynucleotide phosphate terminators to generate high-precision information on gene expression, cell composition, and cell–cell interactions. Image Credit: Tokyo University of Science.
With the development of single-cell RNA sequencing (scRNA-seq), it is now possible to analyze the internal workings of hundreds of cells at once, revolutionizing the areas of biology and medicine. The typical template-switching reaction, which is utilized to make a double-stranded DNA that “complements” the single-stranded RNA and replicates it millions of times, and potential errors in detecting the composition of the cells are the limitations of scRNA-seq approaches.
A novel and better method for scRNA-seq has just been created by a research team from Japan, led by Assistant Professor Shigeyuki Shichino and Professor Kouji Matsushima of Tokyo University of Science. Compared to current, widely-used methods, the new technique, terminator-assisted solid-phase cDNA amplification, and sequencing (TAS-Seq) employ less complicated materials and equipment to produce higher-precision scRNA-seq data.
Our technique, TAS-Seq, combines genetic detection sensitivity, robustness of reaction efficiency, and accuracy of cellular composition to enable us to capture important cellular information.”
Shigeyuki Shichino, Assistant Professor, Research Institute for Biomedical Sciences, Tokyo University of Science
On June 27th, 2022, the work was published in Communications Biology. Associate Professor Satoshi Ueha from Tokyo University of Science, Professor Taka-aki Sato from the University of Tsukuba, and Professor Shinichi Hashimoto from the Wakayama Medical University were also members of the study team.
Terminal transferase is a template-independent enzyme used by TAS-Seq to amp up cDNA (TdT). However, TdT is challenging to manage. Dideoxynucleotide phosphate (ddNTP) was used by the study team as a “terminator” for the cDNA amplification step to overcome this difficulty.
ddNTP spike-in, specifically dideoxycytidine phosphate (ddCTP), stops the excessive extension of polyN-tail by TdT in a stochastic manner, and greatly reduces the technical difficulties of the TdT reaction.”
Shigeyuki Shichino, Assistant Professor, Research Institute for Biomedical Sciences, Tokyo University of Science
The nanowell/bead-based scRNA-seq technology used by TAS-Seq also enables the separation of single cells from tissue samples, reducing cell sampling bias and enhancing the precision of cell composition data.
Using samples of murine and human lung tissue, the study team next confirmed the effectiveness of TAS-Seq and evaluated it against two popular, contemporary scRNA-seq methodologies, 10X Chromium V2 and Smart-seq2. When compared to the most popular scRNA-seq technologies, they discovered that TAS-Seq could not only identify more genes overall but also find more highly variable genes.
We found that TAS-Seq may outperform 10X Chromium V2 and Smart-seq2 in terms of gene detection sensitivity and gene drop-out rates, indicating that TAS-Seq might be one of the most sensitive high-throughput scRNA methods. We can detect genes across a wide range of expression levels more uniformly and also detect growth factor and interleukin genes more robustly.”
Shigeyuki Shichino, Assistant Professor, Research Institute for Biomedical Sciences, Tokyo University of Science
The new technique also has the benefit of TAS-Seq being less prone to batch effects. The fact that TAS-Seq data and flow-cytometric data from the tissue samples had a strong correlation further suggests that TAS-Seq can produce extremely precise cell composition data.
Future, according to Assistant Professor Shichino says, “We have already completed development of TAS-Seq2, an improved, extensively-optimized version of TAS-Seq. TAS-Seq2 has 1.5 to 2 times more sensitive gene detection in mouse spleen cells.” To offer scRNA-seq services utilizing TAS-Seq and TAS-Seq2, the research team has also founded ImmunoGenetics, a venture firm from the Tokyo University of Science.
For scientists working in biology and medicine, scRNA-seq is a crucial tool. The creation of TAS-Seq and TAS-Seq2 will progress the study of “spatial transcriptomics,” which also depends on solid-phase cDNA synthesis, and lead to the identification of novel therapeutic targets for illnesses. Additionally, it will accelerate the advancement of single-cell omics technologies, advancing the comprehension of the biological underpinnings of disease onset and progression.
TAS-Seq: A Novel Technique for Highly Sensitive and Robust Single-Cell RNA Sequencing
The novel terminator-assisted solid-phase complementary DNA amplification and sequencing (TAS-Seq) method provides high-precision data on gene expression. Video Credit: Tokyo University of Science.
Source:
Journal reference:
Shichino, S., et al. (2022) TAS-Seq is a robust and sensitive amplification method for bead-based scRNA-seq. Communications Biology. doi.org/10.1038/s42003-022-03536-0.