In the process of protein synthesis, a peptide bond is formed between two amino acids that are bound to two different transfer RNAs (tRNAs). Researchers have long been perplexed as to how these tRNAs evolved to be so near to one another on the ribosome.
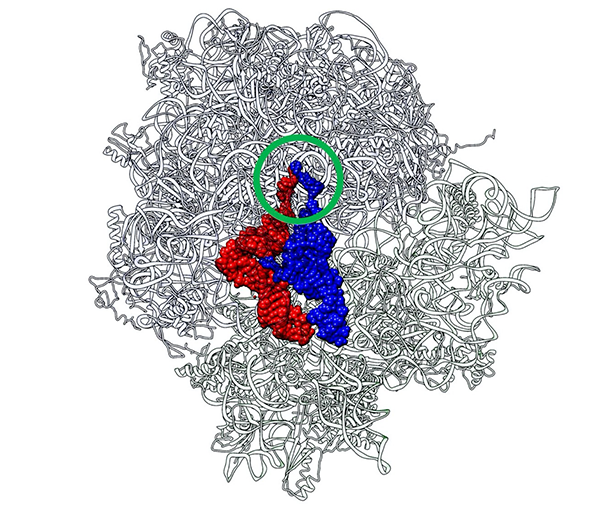 Two CCA termini (circled in green) of P-site tRNA (red) and A-site tRNA (blue) approach in close proximity just like a “Goromaru pose”. The figure was drawn using a coordinate file with PDB ID: 4V5D. Image Credit: Koji Tamura from Tokyo University of Science
Two CCA termini (circled in green) of P-site tRNA (red) and A-site tRNA (blue) approach in close proximity just like a “Goromaru pose”. The figure was drawn using a coordinate file with PDB ID: 4V5D. Image Credit: Koji Tamura from Tokyo University of Science
Researchers describe how tRNA-like components serve as scaffolds for the creation of peptide bonds between amino acid-bound “RNA minihelices,” which are half tRNA-like molecules, in a recent study.
Through the translation process, the genetic information carried in DNA is “decoded” to generate proteins. This entails the establishment of peptide bonds between amino acids attached to transfer RNA (tRNA) molecules that slide over the ribosome in close vicinity to one another, elongating the peptide chain, which then endures conformational variation to create a protein.
In regards to the codon-dependent aminoacyl-tRNA identification in the small ribosomal subunit, peptide bond generation in the major ribosomal subunit happens in a non-amino acid particular manner at the peptidyl transferase center (PTC). This non-specificity suggests that the big subunit developed before the tiny subunit, which interacts with mRNA and tRNA in a more specific manner.
Image Credit: Alila Medical Media/Shutterstock.com
Even though the history of PTC development has been well described, very little is understood about how ribosomes evolved into functional entities and became an essential part of protein synthesis.
The concept that tRNAs require the assistance of a “scaffold” to form a peptide bond, which orients them for engagement via 3’-CCA sequences on their acceptor’s arms, has long-baffled scientists. It would be fascinating to learn more about what that scaffold is and how it works.
Professor Koji Tamura and his team of researchers from the Tokyo University of Science attempted to explore this question from the perspective of biological evolution continuity. Their research, which was published online on April 12, 2022, in the journal Life, Volume 12, Issue 4, gives light on the evolutionary component of protein translation.
Their findings are crucial in proving the hypothesis regarding the PTC’s genesis and evolution, which has revolutionized the way people think about ribosomes and tRNA today.
After studying the crystal structure of the 70S ribosome-tRNA complex from Thermus thermophilus, a bacterium commonly employed in genetics research, the idea developed. Like a rugby player’s index fingers in the “Goromaru pose,” the peptidyl (P-) and aminoacyl (A-) regions of the tRNAs are positioned to put the CCA termini in close contact.
There was a certain entity that served as a scaffold for maintaining this proximity, and it most likely stemmed from the primordial PTC.”
Koji Tamura, Professor, Tokyo University of Science
Since a historical component was anticipated, the researchers chose to explore primordial tRNA, also known as “RNA minihelix.”
In the appearance of a ribosomal RNA segment, researchers sought to create a peptide connection between two alanine-specific minihelices. The peptide bond was created utilizing P1c2, a 70-nucleotide-long ribosomal segment, as an RNA scaffold! They then inserted a terminal amino acid segment (with the sequence UGGU) into the P1c2 (P1c2UGGU).
This boosted the capacity to make peptide bonds by 4.2 times that of the original, as per mass spectrometry studies! A scaffold of dimerized P1c2UGGU promoted the establishment of peptide bonds between two alanine residues.
The scaffold’s UGGU sequence connected with the minihelix’s matching 3’-terminal ACCA, bringing the two amino acids close enough to form a peptide bond. Dr Ada Yonath and her team recently proved that identical, conserved PTC areas might accelerate the creation of peptide bonds with manmade analog molecules, while Professor Tamura’s group demonstrated that an aminoacylated RNA could also be a substrate.
The data strongly suggest that minihelices may bind to the primordial PTC. Therefore, what do these findings say about ribosome evolution?
Functional interactions between the CCA of tRNA and PTC could have been ‘revised’ in the process of evolution. Although current ribosomes do not have a contiguous sequence like UGGU, their interactions are ‘conceptually’ similar to the effects seen in our study. It is plausible that minihelices eventually evolved into tRNA using, for example, kissing-loop interactions between two minihelix-like RNA molecules.”
Koji Tamura, Professor, Tokyo University of Science
“These minihelix-like molecules, which form a part of the scaffold for peptide bond formation, may have not only contributed to the evolution of what is currently the PTC but also formed tRNA molecules,” Tamura added.
The research’s potential developments, which have opened up interesting paths in evolutionary RNA biology, are numerous. Presented with a metabolic conundrum (the fact that amino acids are used to make DNA and RNA), it is possible to look at the idea of “peptide nucleic acids” as genetic material precursors. The findings are remarkable, and they will aid researchers in deciphering molecular processes that have baffled scientists for years.
Source:
Journal reference:
Kawabata, M., et al. (2022) Peptide Bond Formation between Aminoacyl-Minihelices by a Scaffold Derived from the Peptidyl Transferase Center. Life. doi.org/10.3390/life12040573.