Healing debilitating genetic diseases is one of modern medicine’s most difficult challenges. The advancement of CRISPR systems and advances in genetics research over the last decade have given patients and their families a fresh start; however, the safety of these new techniques remains a major concern.
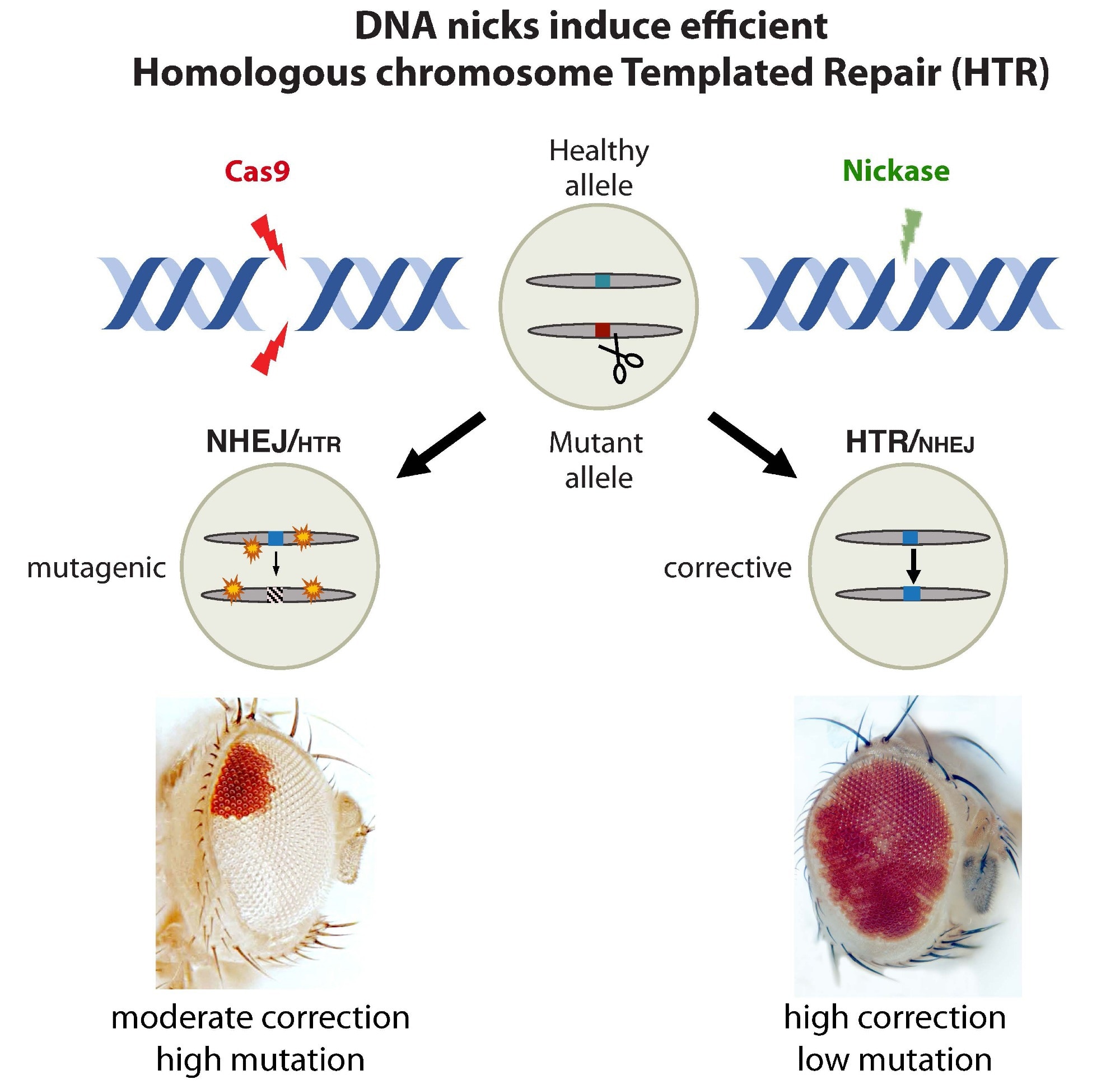 Restorative gene editing using sequences from the counterpart chromosome: The standard CRISPR enzyme Cas9 offers the ability to make repairs but also potentially results in unintended mutations (mutagenic events) at the targeted site and possibly elsewhere in the genome (left). In contrast, the nickase enzyme results in more efficient gene correction and no mutagenic events (right). Image Credit: University of California San Diego
Restorative gene editing using sequences from the counterpart chromosome: The standard CRISPR enzyme Cas9 offers the ability to make repairs but also potentially results in unintended mutations (mutagenic events) at the targeted site and possibly elsewhere in the genome (left). In contrast, the nickase enzyme results in more efficient gene correction and no mutagenic events (right). Image Credit: University of California San Diego
A group of biologists from the University of California, San Diego, led by postdoctoral scholar Sitara Roy, specialist Annabel Guichard, and Professor Ethan Bier, presents an innovative, safe, and secure strategy to correct genetic defects in the journal Science Advances on July 1st, 2022.
Their tactic, which employs natural DNA repair machinery, lays the groundwork for novel gene therapy methods with the possibility to heal a wide range of genetic diseases.
Many people with genetic disorders have unique mutations in the two copies of genes inherited from their parents. This means that a mutation on one chromosome will frequently have a functional sequence counterpart on the other. To take advantage of this fact, the scientists used CRISPR genetic editing features.
The healthy variant can be used by the cell’s repair machinery to correct the defective mutation after cutting the mutant DNA. Remarkably, this can be achieved even more efficiently by a simple harmless nick.”
Annabel Guichard, Study Senior Author and Specialist, University of California, San Diego
Scientists have created mutants in fruit flies that allow them to see such “homologous chromosome-templated repair,” or HTR, by producing pigments in their eyes. At first, such mutants had completely white eyes.
The same flies, however, showcased large red patches across their eyes when CRISPR components (a guide RNA plus Cas9) were demonstrated, indicating that the cell’s DNA repair machinery had achieved overturning the mutation using functional DNA from the other chromosome.
Researchers then put their new system to the test with Cas9 variants recognized as “nickases,” which target only one strand of DNA rather than both. Astonishingly, the researchers noted that such nicks resulted in the high-level restoration of red eye color that was nearly equal to that of normal (non-mutated) healthy flies.
They discovered that the nickase has a 50–70% repair success rate, compared to only 20–30% for dual-strand cutting Cas9, which also creates frequent mutations and targets other places throughout the genome (so-called off-target mutations).
“I could not believe how well the nickase worked—it was completely unanticipated,” said Roy, the lead author of the study. According to the researchers, the new system’s adaptability could serve as a model for correcting genetic mutations in mammals.
We don’t know yet how this process will translate to human cells and if we can apply it to any gene. Some adjustment may be needed to obtain efficient HTR for disease-causing mutations carried by human chromosomes.”
Annabel Guichard, Study Senior Author and Specialist, University of California, San Diego
The new study builds on the group’s past efforts in precision editing with “allelic-drives,” which use a guide RNA to direct the CRISPR system to cut unwanted variants of a gene and replace them with an ideal copy of the gene.
The team’s study is notable for the fact that their nickase-based system results in far fewer on- and off-target mutations than more conventional Cas9-based CRISPR edits. They also claim that a slow, continuous deployment of nickase components over many days may be more advantageous than a single delivery.
“Another notable advantage of this approach is its simplicity,” said Bier. “It relies on very few components and DNA nicks are ‘soft,’ unlike Cas9, which produces full DNA breaks often accompanied by mutations.”
“If the frequency of such events could be increased either by promoting interhomolog pairing or by optimizing nick-specific repair processes, such strategies could be harnessed to correct numerous dominant or trans-heterozygous disease-causing mutations,” said Roy.