Srrm3 is a master regulator gene essential for the growth of photoreceptors, cells in the rear of the retina that catch and process light and transmit signals to the brain that allow vision, according to research, from the Centre for Genomic Regulation (CRG) in Barcelona. Zebrafish with the gene knocked off had severely impaired vision.
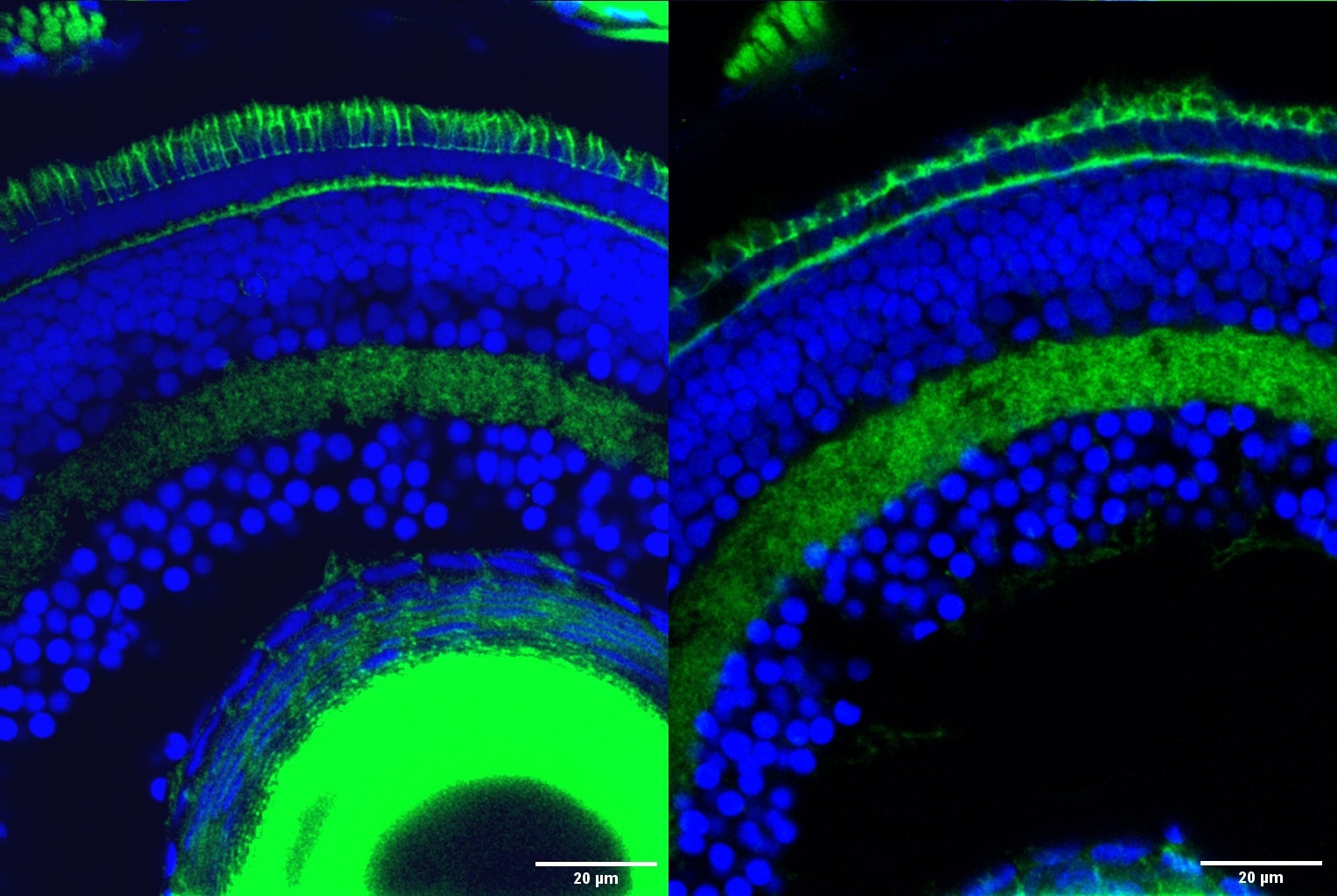 Retinal cells in zebrafish with the outer segment, the part of photoreceptor cells responsible for transforming light into nerve signals that enable vision, stained in green at the top of the image. The outer segment is significantly degraded in retinal cells with the Srrm3 gene knocked out (right) compared to normal retinal cells (left). Image Credit: Ludovica Ciampi/CRG/Proceedings of the National Academy of Sciences
Retinal cells in zebrafish with the outer segment, the part of photoreceptor cells responsible for transforming light into nerve signals that enable vision, stained in green at the top of the image. The outer segment is significantly degraded in retinal cells with the Srrm3 gene knocked out (right) compared to normal retinal cells (left). Image Credit: Ludovica Ciampi/CRG/Proceedings of the National Academy of Sciences
According to the study, Srrm3 controls alternative splicing in vertebrates, a process that enables cells to produce multiple types of proteins from a single gene and is particularly prevalent in neural cells. Misregulation of alternative splicing can have a terrible effect on a person’s health, such as in cases of cancer or neurological conditions.
Researchers discovered that Srrm3 primarily controls the splicing of microexons, little DNA segments that are only 3-27 letters long. Despite their small size, it has been demonstrated that microexon control is crucial for protein and cellular function.
Numerous distinct microexons that are primarily found in photoreceptors but absent from other neurons were discovered by the researchers.
A significant number of these microexons have an impact on the activity of about 70 genes crucial for the growth of a photoreceptor’s outer segment, which is the area of the cell that absorbs light. The results are published in the journal Proceedings of the National Academy of Sciences.
The study indicates a new level of cellular specialization needed for retinal cells’ distinctive cellular form and function, one of the body’s most sophisticated and specialized cells. Due to their intricacy, retinal cells are dependent on numerous distinct genes for their development—any one of these genes could be mutated to produce disease and cause blindness.
Retinitis pigmentosa, a hereditary condition for which the underlying molecular pathways are poorly understood, is one of the most frequent causes of inherited eyesight loss.
About 40–50% of retinitis pigmentosa cases are unexplained, meaning they involve mutations in unidentified genes. The study’s authors to determine whether Srrm3 or the microexons concerned can explain some of these situations will conduct future research.
The Srrm3 gene has neither been associated with the development of photoreceptor cells nor with the pathogenesis of retinal diseases before. We are already exploring the gene’s role in patients without a genetic diagnosis. If we find cases with mutations in this specific gene, or on any retinal microexons, it could lead to potential new therapeutic strategies to manage the condition.”
Ludovica Ciampi, Study First Author and PhD Student, Centre for Genomic Regulation
For finding new therapeutic targets, microexon regulation in certain cell types must be understood, claims ICREA Research Professor Manuel Irimia.
“Photoreceptors have unique properties thanks to the regulation of alternative splicing and microexons. This helps make the cell more specialized but also perhaps more susceptible to genetic diseases. Modulating splicing activity is now possible, so the more intricate biology we uncover, the more likely we are to find therapeutic targets to treat retinal diseases,” concludes Dr Irimia.
Source:
Journal reference:
Ciampi, L., et al. (2022) Specialization of the photoreceptor transcriptome by Srrm3-dependent microexons is required for outer segment maintenance and vision. Proceedings of the National Academy of Sciences. doi.org/10.1073/pnas.2117090119.