Filamentous fungi have long been a close ally of sake brewers, but they may soon become associates of environmentalists as well. Scientists from Osaka Metropolitan University discovered the regulatory mechanisms of enzyme synthesis in a filamentous fungus, allowing for the effective degradation of plant biomass, an alternative energy resource to petroleum.
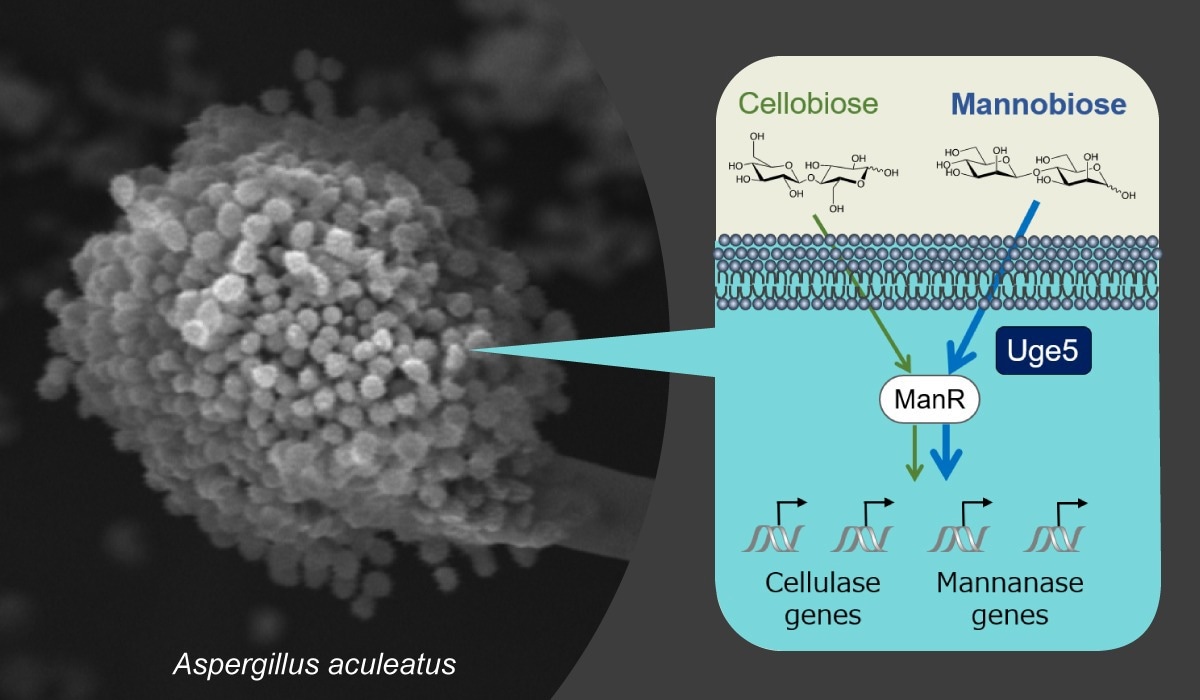 A scanning electron microscopy image of the filamentous fungus Aspergillus aculeatus and a gene regulation model in A. aculeatus. Image Credit: Osaka Metropolitan University
A scanning electron microscopy image of the filamentous fungus Aspergillus aculeatus and a gene regulation model in A. aculeatus. Image Credit: Osaka Metropolitan University
Filamentous fungi (molds) are microbes that have been used for decades in the fermentation of sake, soy sauce, cheese, and a variety of other products. This fermentation is an excellent illustration of the industrial application of filamentous fungi's capacity to release a variety of enzymes in great quantities.
Plant biomass is currently gaining popularity as a substitute for petroleum, which will soon be depleted. Because rigid plant cell walls are made up of a variety of aromatics and polysaccharides, their breakdown necessitates the use of a diverse set of enzymes. As a result, research has been performed to use filamentous fungi as a major supply of enzymes for plant biomass breakdown.
A team of researchers headed by Associate Professor Shuji Tani from Osaka Metropolitan University’s Graduate School of Agriculture investigated the regulatory mechanisms of carbohydrate-hydrolyzing enzyme production in the filamentous fungus Aspergillus aculeatus, which produces enzymes with an outstanding ability to degrade plant biomass.
Uridine diphosphate (UDP)-glucose 4-epimerase (Uge5) is a widely used galactose metabolism enzyme. Uge5 does, however, influence the expression of degrading enzyme genes in A. aculeatus, according to the researchers. This is the first report of Uge5’s activities in selective gene expression in filamentous fungi in response to distinct types of inducing sugars.
These observations address the present technological problem of developing a complete high-production technique for different enzymes in filamentous fungi.
We constructed and screened a library containing approximately 9,000 gene-disrupted strains of Aspergillus aculeatus, and identified Uge5 as a novel regulatory factor that regulates the production of carbohydrate-hydrolyzing enzymes. The discovery of this new function took us by surprise. We plan to continue our research to elucidate phenomena that existing knowledge cannot explain.”
Shuji Tani, Associate Professor, Graduate School of Agriculture, Osaka Metropolitan University
Source:
Journal reference:
Kuga, M., et al. (2023) A new function of a putative UDP-glucose 4-epimerase on the expression of glycoside hydrolase genes in Aspergillus aculeatus. Applied Microbiology and Biotechnology. doi.org/10.1007/s00253-022-12337-8.