Pep-tRNAs, nascent polypeptides inside the ribosome that are covalently connected to transfer RNA, are engaged in a variety of cell processes, including gene expression, according to advances in molecular biology.
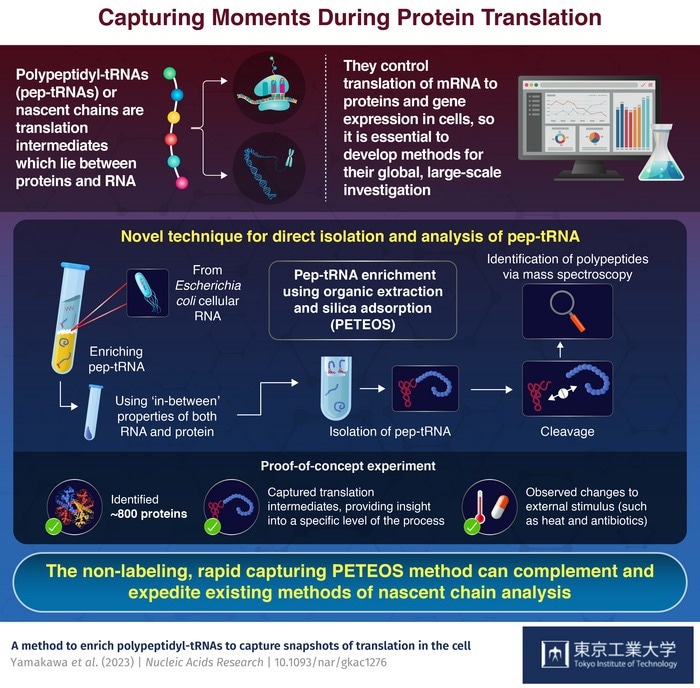
Image Credit: Tokyo Tech
All proteins start as pep-tRNAs, and research into these translational intermediates is essential since they have characteristics of both RNA and protein and can aid in understanding the details of the translation.
Translational regulation is extremely quick and encompasses initiation, elongation, and elongation halting according to inputs and/or stressors. Therefore, developing an appropriate approach to process pep-tRNAs in large quantities is necessary to get deeper insights into the process of translation.
The introduction of molecular tools to study cellular translation has been sparked by these subtleties.
The two main methods utilized to study cellular translation at the moment involve radically different techniques. The first, ribosome profiling, is an effective deep-sequencing technique that tracks translation by focusing on mRNA segments that have been protected by the ribosome as a result of RNase digestion.
Unfortunately, this method is time-consuming, costly, and data-intensive, and it is essentially unable to collect pep-tRNAs. As an alternative, polypeptides must be marked with unique amino acids utilizing liquid chromatography-tandem mass spectrometry (LC-MS/MS).
However, since pep-tRNAs are not specifically targeted, such proteomics techniques take a lot of time and, more significantly, do not accurately reflect the cell’s translation elongation state.
A team of researchers from the Tokyo Institute of Technology (Tokyo Tech), led by Prof. Hideki Taguchi, created PETEOS to overcome these drawbacks in “pep-tRNA-ome” technology. Nucleic Acids Research has published the findings.
This non-labeling methodology is unique in that it specifically enriches and rapidly captures pep-tRNAs while still complimenting conventional translatome analyses platforms.”
Hideki Taguchi, Professor, Cell Biology Center, Institute of Innovative Research, Tokyo Institute of Technology
Four processes make up PETEOS. RNA is extracted from enriched samples using an organic solvent before being isolated on a silica column. Next, a combination of high pH and temperature is used to separate the target polypeptides from their ribonucleotides in the isolated pep-tRNAs.
The released polypeptides are then cleaved and digested by an enzyme. Lastly, shotgun LC-MS/MS proteomics is used to identify these polypeptides.
Taguchi added, “As a proof-of-concept, we were able to identify nearly 800 E. coli proteins that were derived from pep-tRNA using PETEOS. Importantly, these proteins were N-termini enriched which underscores that the method really does capture the intermediate pep-tRNA pool during translation.”
Additionally, the group demonstrated that PETEOS could record “screenshots” of the nascentome, or the collection of nascent pep-tRNAs that are always present when E. coli cells were exposed to heat shock and antibiotic treatments.
In terms of the ability to record acute protein expression under heat stress and the reorganization of the nascentome after exposure to various antibiotics, PETEOS fared better than traditional approaches.
There are certain drawbacks to PETEOS, such as the difficulty in employing LC-MS/MS to analyze low-quantity pep-tRNAs, the acidic phenol/chloroform step’s limited quantitative capability, the lack of codon-level precision, and the challenge in capturing the C-terminal end of the developing polypeptide.
The effectiveness of this procedure’s C-terminal peptide enrichment can be increased, albeit some of these problems apply to traditional methods as well.
The team believes there will be numerous opportunities for protocol optimization in the future.
Taguchi concluded, “Despite the limitations, we are confident PETEOS has potential as it enables analyses that current translatome methods simply cannot. Most importantly, it can be applied to any organism!”
The understanding of protein synthesis could be furthered with additional study in this area.
Source:
Journal reference:
Yamakawa, A., et al. (2023). A method to enrich polypeptidyl-tRNAs to capture snapshots of translation in the cell. Nucleic Acids Research. doi.org/10.1093/nar/gkac1276