In Jennifer Hines’ lab, a group of graduate and undergraduate chemistry students recently discovered a new family of compounds that can target RNA and interfere with its function. In the future, RNA-targeted treatments to treat cancer and metabolic diseases, together with bacterial and viral infections, could be developed using the chemical scaffold found in this research.
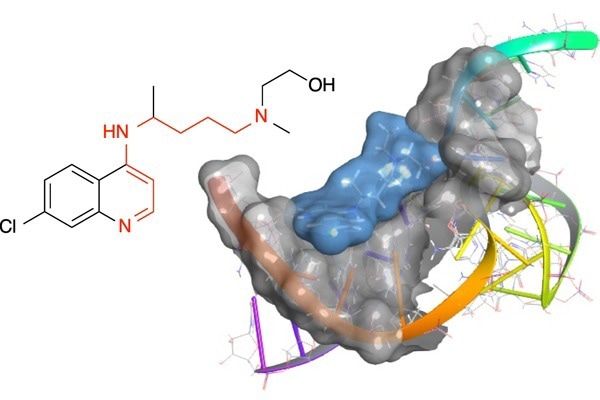 4-Aminoquinolines inhibit the function of the bacterial T-box riboswitch RNA. Image Credit: Jennifer Hines
4-Aminoquinolines inhibit the function of the bacterial T-box riboswitch RNA. Image Credit: Jennifer Hines
Chemically similar to DNA, RNA also regulates how much a living cell follows the instructions in the DNA. The fact that RNA plays such a crucial regulatory role in cellular activity makes it such a desirable target.
Trying to target RNA is at the forefront of medicinal chemistry research with enormous potential for treating diseases. However, there are relatively few compounds known to directly modulate RNA activity which makes it challenging to design new RNA-targeted therapeutics.”
Jennifer Hines, Professor, Chemistry and Biochemistry, College of Arts and Sciences, Ohio University
The Hines group discovered that 4-aminoquinolines bind to stem-loop II motif RNA and limit the activity of the bacterial T-box riboswitch RNA (an RNA structure found in the virus causing the COVID-19 pandemic).
Hines added, “The compounds bind these RNA structures in very specific sites, making them good starting scaffolds for designing specific therapeutics. What was so surprising about this discovery is that the likelihood for RNA binding was hiding in plain sight within the 4-aminoquinoline structure, but no one had identified it before.”
She further stated, “Our research determined that 4-aminoquinolines have distinct activities and chemical features very similar to polyamines which are natural compounds in the cell that modulate RNA function.”
The study’s co-first authors were Mason Myers, who earned a B.S. in Chemistry from the Honors Tutorial College (HTC) in OHIO in 2021 and graduated from the university with honors, and graduate student Md. Ismail Hossain. At the University of Michigan, Myers is now a doctoral student studying chemistry and biochemistry.
In addition to Hannah Boesger, an undergraduate student at HTC who achieved a B.S. in Neuroscience in 2022, other writers included graduate students Danushika Herath and Ali Aldhumani. Boesger is currently a PhD student in medicinal chemistry at the University of Michigan’s College of Pharmacy.
Hines added, “As part of a comprehensive RNA-targeted drug discovery project, we have been focused on investigating ligand-RNA binding interactions involving larger, more dynamic RNA structural motifs for more than 20 years. This experience is what enabled my group to so quickly respond, when the pandemic began, to investigate targeting the viral stem-loop II motif RNA first virtually via computational studies and then in the lab.”
In their RNA-targeted drug discovery studies, the Hines group combines spectroscopic methods (fluorescence, UV-Vis, NMR), biochemical/biophysical methods (gel electrophoresis, isothermal titration calorimetry), and computational methods (docking, molecular dynamics simulations, quantitative structure activity calculations, bioinformatics).
“It was in this earlier study where we first noticed the 4-aminoquinolines, but not enough was known about the function of the stem-loop II motif RNA to discern what the compounds might be doing,” further added Hines.
Hines concluded, “Consequently, we shifted to exploring the functional effect of these compounds on the T-box riboswitch RNA, which regulates gene expression in bacteria. In these riboswitch studies, we found that the compound’s inhibitory effect was dose-dependent in a manner very similar to the dose-dependency of polyamines, a class of compounds that normally bind RNA in the cell. It was in puzzling out why this might be the case when I noticed the structural similarity between the two classes of compounds.”
Source:
Journal reference:
Hossain, M. I., et al. (2023). 4-Aminoquinolines modulate RNA structure and function: Pharmacophore implications of a conformationally restricted polyamine. Biochemical and Biophysical Research Communications. doi.org/10.1016/j.bbrc.2022.12.080