Each gene in the human DNA has a start and an endpoint. It is critical to correctly define the gene's extremities to make functional proteins. Much study has been conducted to identify what determines when, where, and at which place on the DNA a gene “starts.”
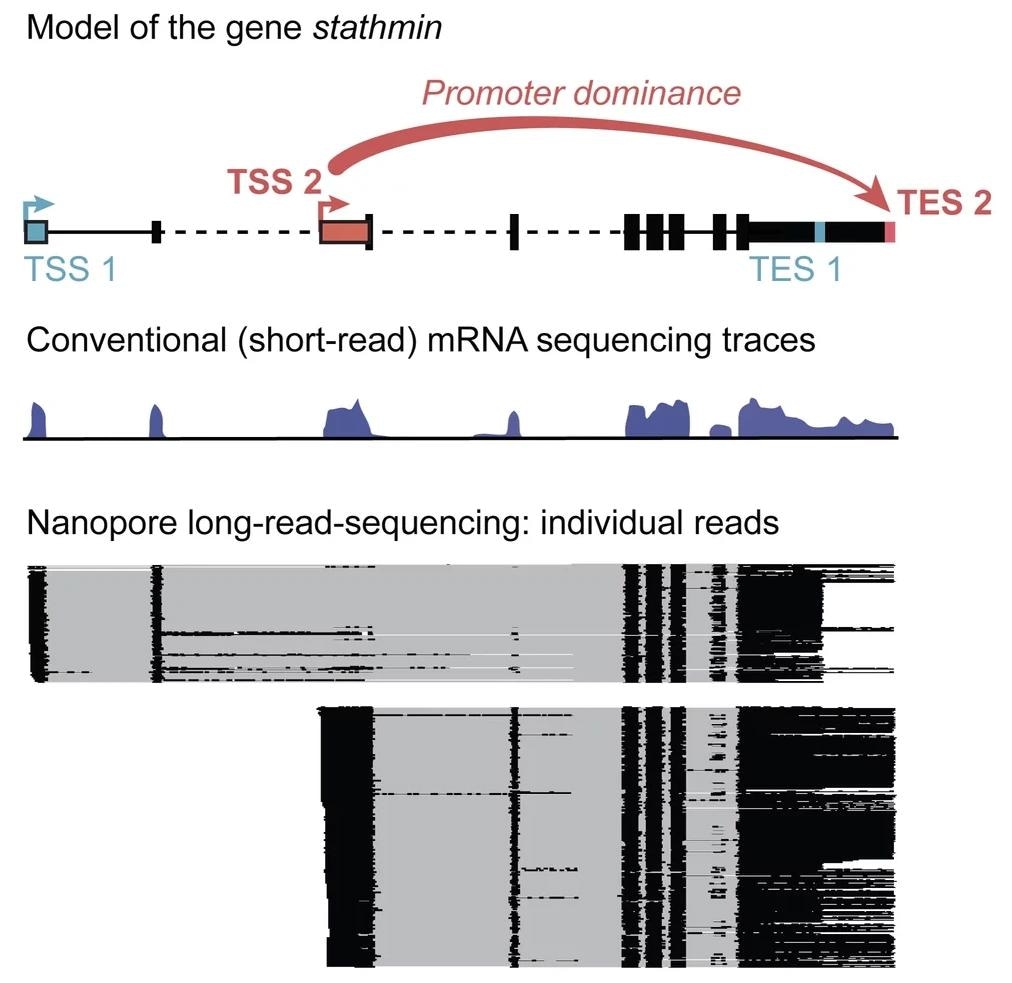 Example of a gene that contains two possible start sites (TSS), and two possible end sites (TES). Black boxes in the gene model show sequences that will be translated into protein. With conventional short-read mRNA sequencing, in which signal represents the accumulation of reads, it is not possible to distinguish different molecules. In long-read sequencing, each horizontal line represents one mRNA molecule from beginning to end. For the gene stathmin, we can see that TSS1 predetermines ending at TES1, and TSS2 will lead to TES2. Image Credit: MPI of Immunobiology and Epigenetics/ Hilgers
Example of a gene that contains two possible start sites (TSS), and two possible end sites (TES). Black boxes in the gene model show sequences that will be translated into protein. With conventional short-read mRNA sequencing, in which signal represents the accumulation of reads, it is not possible to distinguish different molecules. In long-read sequencing, each horizontal line represents one mRNA molecule from beginning to end. For the gene stathmin, we can see that TSS1 predetermines ending at TES1, and TSS2 will lead to TES2. Image Credit: MPI of Immunobiology and Epigenetics/ Hilgers
But where a gene terminates is a different story: transcription termination sites are believed to be selected by downstream elements and external influences.
In their most recent research, scientists from the Max Planck Institute of Immunobiology and Epigenetics in Freiburg, Germany, discovered the start site of transcription controls the final site of transcription for the majority of human genes.
This process, which predetermines mRNA end sites at the very beginning of transcription and is highly conserved across species, is essential for cell identity and functionality.
An organism’s cells all have the same DNA sequence. The assortment of genes that will be activated in a certain location at a specific time dictates the identity and function of particular cells and tissues.
The messenger RNA (mRNA) molecules produced by these active genes, which are transcribed from the DNA template, will encode the proteins required for cellular activity.
A sophisticated molecular mechanism starts converting DNA sequences into mRNA at specific places known as promoters. It is interesting to note that most genes have numerous places where transcription might begin or terminate. This implies that the mRNAs for each gene might vary based on the start or termination position.
One gene can be expressed in several distinct ways, significantly enhancing the genome’s variety and utility. It simultaneously makes studying the genome more complicated.
RNA Snapshots from Beginning to End
Researchers at the Max Planck Institute of Immunobiology and Epigenetics in Freiburg were interested in learning how many start and end sites each gene employs, what combinations they utilize, and whether those combinations vary depending on the environment.
The technical problem to answer this question is that we have to “read” each and every mRNA molecule from all genes from the very beginning to the very end. This a humongous task that has not been undertaken before.”
Valérie Hilgers, Research Group Leader, Max Planck Institute of Immunobiology and Epigenetic
Hilgers is also a member of the Cluster of Excellence CIBSS—Centre for Integrative Biological Signalling Studies at the University of Freiburg.
The researchers read each individual mRNA using modified next-generation sequencing techniques. Each mRNA is divided into smaller pieces for traditional short-read sequencing, which results in the “read” after sequencing. Following that, a continuous sequence is created by piecing the reads together using bioinformatic algorithms.
The Hilgers collaborated with the Max Planck Institute’s Deep Sequencing Facility to optimize particular long-read-sequencing technologies for full-length mRNA information of the whole genome in many Drosophila tissues, including the brain.
Long-read sequencing allows for the retrieval of much longer sequencing reads than widely used standard sequencing. However, we even had to optimize this technology and increase the typical read length by several fold to obtain full-length mRNA information in our different model systems.”
Carlos Alfonso-Gonzalez, Study First Author and PhD Student, Max Planck Institute of Immunobiology and Epigenetics
The Hilgers Lab also used cerebral organoids, which are “mini-brains” produced in a dish from induced pluripotent stem cells, as a human model of the nervous system in their research in addition to Drosophila.
Transcription End Sites Are Pre-Determined at Transcription Start
The obtained information, which represents each mRNA at the level of the whole molecule, provides hitherto unheard-of insight into the transcription of specific genes.
Hilgers added, “We realized that far from start sites (TSSs) and end sites (TESs) being randomly combined one to another, we found that often, sites of transcription start are specifically linked to distinct sites of transcription end.”
This connection is causative; for instance, in ovaries, a TSS that is typically only utilized in the brain is intentionally activated, which overrides the natural TES and artificially induces the usage of the brain TES. This demonstrates the crucial part TSS plays in defining the distinctive RNA landscape of each tissue and consequently affecting tissue identity.
One phenomenon, though, stood out.
Alfonso-Gonzalez further stated, “Certain TSSs show unexpected dominance behavior. They overrule conventional signals to end transcription, outcompete other TSSs, and cause the selection of distinct TESs. Accordingly, we named them »dominant promoters.”
The scientists also discovered that certain epigenetic markers were used to direct connections between these dominating promoters and the gene ends that they were linked with.
The findings in Drosophila brain cells were particularly significant since they could be reproduced in human brain organoids, demonstrating that promoter dominance is a conserved, maybe universal, mechanism for controlling the synthesis of useful proteins and the functioning of the cells.
What potential physiological implications could this new mechanism have? The Freiburg researchers found that TSSs and TESs exhibit co-evolution by showing individual nucleotide changes at dominant promoters in the gene start, which were accompanied by changes at the corresponding gene end, over millions of years of evolution between species.
Hilgers concluded, “We interpret this observation as a “push” through evolution, to sustain the interaction between both extremities of the gene, which implies significant importance of these couplings for animal fitness.”
Source:
Journal reference:
Alfonso-Gonzalez, C., et al. (2023). Sites of transcription initiation drive mRNA isoform selection. Cell. doi.org/10.1016/j.cell.2023.04.012