Reviewed by Danielle Ellis, B.Sc.Jun 29 2023
Feng Zhang of the Broad Institute at MIT and Harvard and the McGovern Institute for Brain Research at MIT headed a team of researchers who discovered the first programmable RNA-guided mechanism in eukaryotes, which include fungi, plants, and animals.
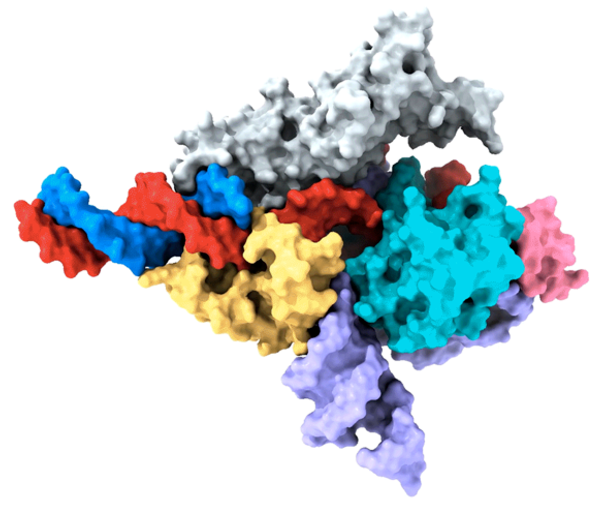 Cryo-EM map of a Fanzor protein in complex with ωRNA and its target DNA. Image Credit: Zhang lab.
Cryo-EM map of a Fanzor protein in complex with ωRNA and its target DNA. Image Credit: Zhang lab.
The team reveals how the system is built on a protein called Fanzor in a study published in Nature. They demonstrated that Fanzor proteins employ RNA as a guide to accurately target DNA and that Fanzors can be reprogrammed to change human cell genomes.
The tiny Fanzor systems may be more easily administered to cells and tissues as therapeutics than CRISPR/Cas systems, and additional modifications to improve targeting effectiveness may make them a valuable new technique for human genome editing.
CRISPR/Cas was found in prokaryotes (bacteria and other single-cell organisms without nuclei), and researchers, including Zhang’s lab, have long speculated about whether analogous systems exist in eukaryotes. The new research shows that RNA-guided DNA-cutting processes exist in all kingdoms of life.
CRISPR-based systems are widely used and powerful because they can be easily reprogrammed to target different sites in the genome. This new system is another way to make precise changes in human cells, complementing the genome editing tools we already have.”
Feng Zhang, Senior Study Author and Core Institute Member, Broad Institute
Zhang is also an investigator at MIT’s McGovern Institute, the James and Patricia Poitras Professor of Neuroscience at MIT, and a Howard Hughes Medical Institute investigator.
Searching the Domains of Life
The Zhang lab's main goal is to create genetic medicines by using systems that can control human cells by specifically targeting genes and processes. “A number of years ago, we started to ask, ‘What is there beyond CRISPR, and are there other RNA-programmable systems out there in nature?’” says Zhang.
Zhang lab members identified OMEGAs, a class of RNA-programmable systems in prokaryotes that are frequently connected with transposable elements, or “jumping genes,” in bacterial genomes and likely gave rise to CRISPR/Cas systems, two years ago. That research also found similarities between prokaryotic OMEGA systems and eukaryotic Fanzor proteins, implying that Fanzor enzymes may also use an RNA-guided mechanism to identify and cut DNA.

Image Credit: nobeastsofierce/Shutterstock.com
The researchers extended their research of RNA-guided systems in the latest study by separating Fanzors from fungi, algae, and amoeba species, as well as a clam known as the Northern Quahog.
The biochemical characterization of the Fanzor proteins was led by co-first author Makoto Saito of the Zhang lab, who demonstrated that they are DNA-cutting endonuclease enzymes that utilize adjacent non-coding RNAs known as ωRNAs to target specific places in the genome. This mechanism has never been discovered in eukaryotes, such as animals.
Unlike CRISPR proteins, Fanzor enzymes are encoded within transposable elements in the eukaryotic genome, and the team’s phylogenetic study implies that Fanzor genes transferred from bacteria to eukaryotes via so-called horizontal gene transfer.
“These OMEGA systems are more ancestral to CRISPR and they are among the most abundant proteins on the planet, so it makes sense that they have been able to hop back and forth between prokaryotes and eukaryotes,” said Saito.
The investigators have found that Fanzor can induce insertions and deletions at selected genomic regions within human cells to investigate its feasibility as a genome editing tool. The Fanzor system was initially discovered to be less effective at snipping DNA than CRISPR/Cas systems, but by methodical engineering, the researchers induced a combination of mutations into the protein that enhanced its activity 10-fold.
Furthermore, unlike several CRISPR systems and the OMEGA protein TnpB, the researchers discovered that a fungal-derived Fanzor protein lacked “collateral activity,” which occurs when an RNA-guided enzyme cleaves its DNA target while also destroying surrounding DNA or RNA. The findings show that Fanzors could be created as effective genome editors.
Peiyu Xu, the co-first author, spearheaded an attempt to examine the molecular structure of the Fanzor/ωRNA complex and show how it latches onto DNA to cut it. Fanzor shares structural similarities with its prokaryotic counterpart, the CRISPR-Cas12 protein, but the contact between the ωRNA and Fanzor’s catalytic domains is more extensive, implying that the ωRNA may play a role in the catalytic reactions.
“We are excited about these structural insights for helping us further engineer and optimize Fanzor for improved efficiency and precision as a genome editor,” Xu notes.
The Fanzor system, like CRISPR-based systems, can be readily reprogrammed to target specific genomic sites, and Zhang believes it has the potential to become a powerful new genome editing method for research and therapeutic applications. The abundance of RNA-guided endonucleases like Fanzors adds to the number of OMEGA systems known across kingdoms of life, implying that there are more to be discovered.
Nature is amazing. There’s so much diversity. There are probably more RNA-programmable systems out there, and we're continuing to explore and will hopefully discover more.”
Feng Zhang, Senior Study Author and Core Institute Member, Broad Institute
Source:
Journal reference:
Saito, M., et al. (2023). Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature. doi.org/10.1038/s41586-023-06356-2.