Reviewed by Danielle Ellis, B.Sc.Dec 11 2023
Novel insights into antibody aggregation are anticipated to pave the way for fresh avenues in both research and therapeutic applications.
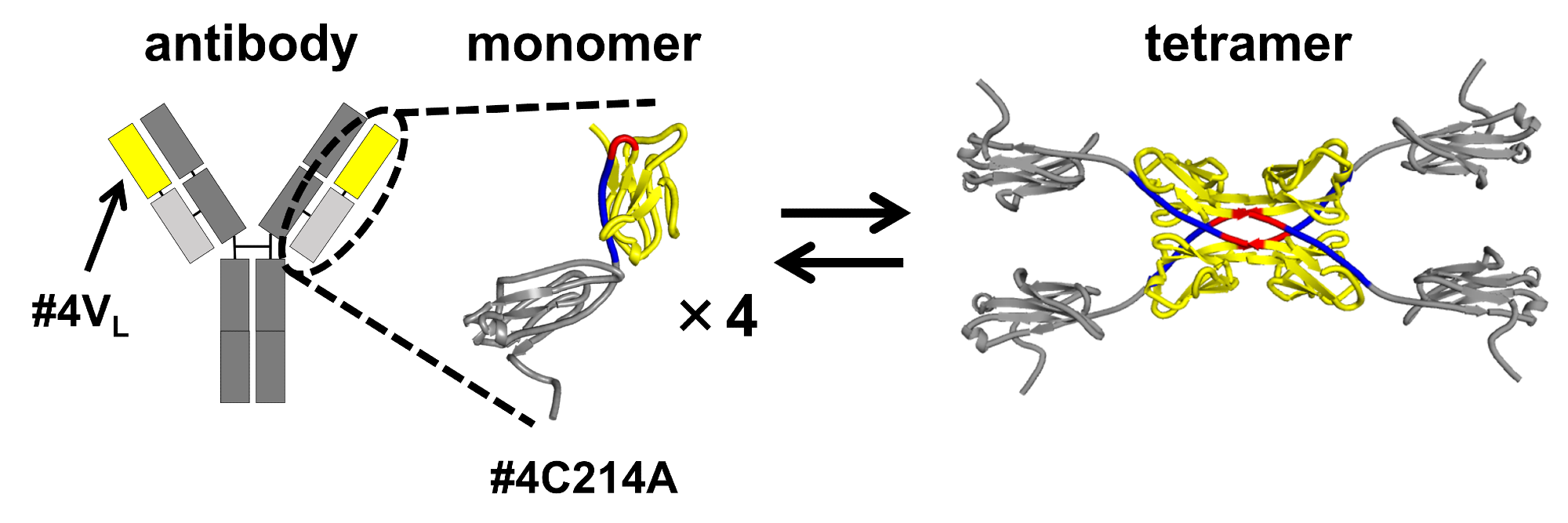 The schematic structures resulting from 3d domain swapping of the variable region #4vl (yellow) in the light chain #4c214a (yellow and light gray) of an antibody. The light chain of the antibody was found to maintain an equilibrium between monomeric and tetrameric states. Image Credit: Shun Hirota and Takahiro Sakai.
The schematic structures resulting from 3d domain swapping of the variable region #4vl (yellow) in the light chain #4c214a (yellow and light gray) of an antibody. The light chain of the antibody was found to maintain an equilibrium between monomeric and tetrameric states. Image Credit: Shun Hirota and Takahiro Sakai.
Antibodies, also known as immunoglobulins, are Y-shaped proteins designed to identify and neutralize specific pathogens. Their capacity to target precise molecules or cells positions them as promising candidates for future drug development.
Despite their potential, the light chains of antibodies, crucial for recognizing and binding to specific antigens, can misfold and aggregate.
This aggregation leads to amyloidosis, a condition causing complications and tissue dysfunction in the body. In the realm of drug development, antibody aggregation poses a challenge as it can compromise their ability to bind to antigens, thereby reducing their therapeutic efficacy.
One impediment to progress in this field is the lack of detailed structural information regarding antibody aggregation. Ongoing efforts are focused on providing comprehensive reports on aggregate structures and their formation mechanisms to advance antibody drug development.
Image Credit: Mirror-Images/Shutterstock.com
A recent study published in Nature Communications by a team of Japanese researchers, led by Shun Hirota from the Nara Institute of Science and Technology (NAIST), has shed light on the structures formed during antibody aggregation through a process called 3D domain swapping (3D-DS).
This process involves the exchange of a specific region of a protein between two or more molecules of the same protein. While 3D-DS has been observed in various proteins, this study is the first to identify it in antibody light chains.
In their investigation, the researchers used a modified version of the antibody light chain. They replaced a cysteine (Cys) residue, which typically forms a disulfide bond with a heavy chain cysteine, with alanine (Ala).
This modification allowed the team to isolate and study the structures resulting from 3D-DS in the segment of the antibody contributing to antigen binding. The 3D-DS of the antibody light chain involved the formation of dimers (structures consisting of two identical subunits) and tetramers (structures composed of two dimers with four identical subunits).
Our study provides the first report on the atomic-level structure of the 3D-DS phenomenon in an antibody light chain's variable region.”
Shun Hirota, Researcher, Nara Institute of Science and Technology
The size exclusion chromatography of antibody light chain #4C214A demonstrated the presence of both individual monomers and tetramers composed of four subunits.
To identify the region responsible for tetramer formation, researchers divided the antibody light chain into the variable region (located at the tip of the Y-shaped antibody) and the constant region (situated in the middle part of the Y-shaped antibody). Their investigation revealed that the variable region #4VL has the capability to transition between monomeric and tetrameric states.
Subsequent examinations utilizing X-Ray crystallography and thermodynamic simulations unveiled that the driving force behind tetramer formation lies in hydrophobic interactions occurring between two 3D-DS dimers.
In comparison to monomers, tetramers exhibited more rigid β-sheet structures, rendering them less flexible. The creation of 3D-DS tetramers could play a role in preventing protein aggregation by reducing flexibility, and potentially mitigating the formation of insoluble aggregates. However, it is important to note that 3D-DS might also contribute to the aggregation of antibodies.
These findings not only clarify the domain-swapped structure of the antibody light chain but also contribute to controlling antibody quality and advancing the development of future molecular recognition agents and drugs.”
Shun Hirota, Researcher, Nara Institute of Science and Technology
Source:
Journal reference:
Sakai, T., et al. (2023) Structural and thermodynamic insights into antibody light chain tetramer formation through 3D domain swapping. Nature Communications. doi.org/10.1038/s41467-023-43443-4.