The genetic code is composed of just four letters representing the four nucleotides, which are the fundamental building blocks of DNA. Scientists have long contemplated the possibility of expanding this genetic alphabet by generating novel nucleotides in the laboratory.
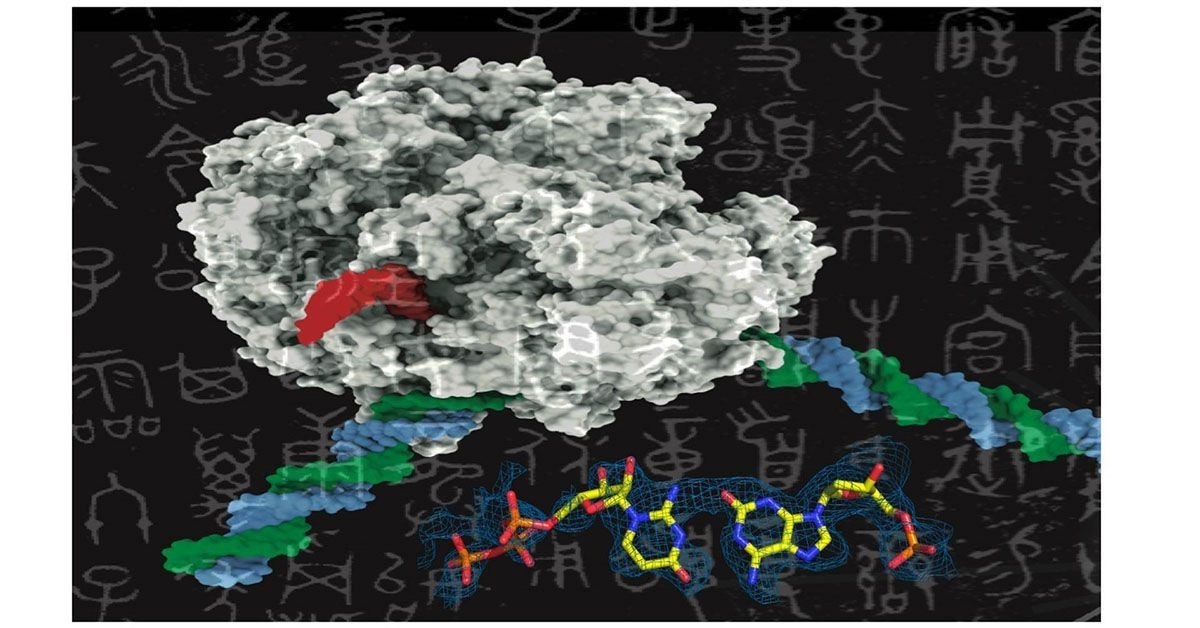 Like adding new letters to an existing language’s alphabet to expand its vocabulary, adding new synthetic nucleotides to the genetic alphabet could expand the possibilities of synthetic biology. This image shows a rendering of RNA polymerase (center) and a synthetic nucleotide (lower right). Image credit: UC San Diego Health Sciences.
Like adding new letters to an existing language’s alphabet to expand its vocabulary, adding new synthetic nucleotides to the genetic alphabet could expand the possibilities of synthetic biology. This image shows a rendering of RNA polymerase (center) and a synthetic nucleotide (lower right). Image credit: UC San Diego Health Sciences.
The feasibility of such innovation hinges on whether cells can effectively identify and utilize these artificial nucleotides for protein synthesis.
In a significant advancement, researchers at the Skaggs School of Pharmacy and Pharmaceutical Sciences at the University of California San Diego have made strides in unlocking the potential of artificial DNA.
Their study revealed that RNA polymerase, a crucial enzyme in protein synthesis, demonstrated the ability to recognize and transcribe an artificial base pair in a manner identical to its interaction with natural base pairs.
These study findings open avenues for scientists to design custom proteins, potentially contributing to developing new medicines.
The study was reported in the journal Nature Communications on December 12th, 2023.
Considering how diverse life on Earth is with just four nucleotides, the possibilities of what could happen if we can add more are enticing. Expanding the genetic code could greatly diversify the range of molecules we can synthesize in the lab and revolutionize how we approach designer proteins as therapeutics.
Dong Wang Ph.D., Study Senior Author and Professor, Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego
Wang collaborated as a co-leader in the study alongside Steven A. Benner, Ph.D., from the Foundation for Applied Molecular Evolution, and Dmitry Lyumkis, Ph.D., from the Salk Institute for Biological Studies.
The DNA molecule is composed of four nucleotides known as adenine (A), thymine (T), guanine (G), and cytosine (C). Within a DNA molecule, these nucleotides engage in base pairs with a distinctive molecular geometry referred to as Watson and Crick geometry. This term pays homage to the scientists who unveiled the double-helix structure of DNA in 1953.
The Watson and Crick base pairs consistently form in two configurations: A-T and C-G. The collective assembly of numerous Watson and Crick base pairs gives rise to the double-helix structure of DNA.
This is a remarkably effective system for encoding biological information, which is why serious mistakes in transcription and translation are relatively rare. As we’ve also learned, we may be able to exploit this system by using synthetic base pairs that exhibit the same geometry.
Dong Wang, Study Senior Author and Professor, Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego
The research employs a modified version of the conventional genetic alphabet known as the Artificially Expanded Genetic Information System (AEGIS), which introduces two additional base pairs. Initially conceived by Benner, AEGIS originated as a NASA-backed initiative aiming to explore the potential development of extraterrestrial life.
By isolating RNA polymerase enzymes from bacteria and subsequently testing their interactions with synthetic base pairs, the study revealed that the AEGIS synthetic base pairs exhibit a geometric structure mirroring the Watson and Crick geometry observed in natural base pairs.
Consequently, the enzymes responsible for DNA transcription cannot discern any distinction between these synthetic base pairs and those naturally occurring.
In biology, structure determines function. By conforming to a similar structure as standard base pairs, our synthetic base pairs can slip in under the radar and be incorporated in the usual transcription process.
Dong Wang, Study Senior Author and Professor, Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego
Beyond broadening the horizons of synthetic biology, the results also lend support to a hypothesis tracing back to the time of Watson and Crick’s groundbreaking discovery. Known as the tautomeric hypothesis, it posits that the conventional four nucleotides can create mismatched pairs as a consequence of tautomerization.
Tautomerization refers to the propensity of nucleotides to oscillate among various structural variants while maintaining the same composition. This phenomenon is believed to be a contributing factor to point mutations, wherein genetic alterations affect only one base pair within a DNA sequence.
Wang stated, “Tautomerization allows nucleotides to come together in pairs when they aren’t usually supposed to. Tautomerization of mispairs has been observed in replication and translation processes, but here we provide the first direct structural evidence that tautomerization also happens during transcription.”
The researchers are now keen to explore whether the observed effect remains consistent when testing other combinations of synthetic base pairs and cellular enzymes.
Wang stated, “We are excited to assemble a multidisciplinary collaborative team with Steve and Dmitry that allow us to tackle the molecular basis of transcription on expanded alphabet. There could be many other possibilities for new letters besides what we’ve tested here, but we need to do more work to figure out how far we can take it.”
Source:
Journal reference:
Oh, J., et al. (2023) A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase. Nature Communications. doi.org/10.1038/s41467-023-43735-9.