Reviewed by Danielle Ellis, B.Sc.Jan 17 2024
It has long been a mystery how the complex molecular machinery of life sprang from modest beginnings. Numerous lines of evidence suggest that life originated in a primordial "RNA world" where an "RNA copy machine," or replicase, began copying itself and other RNA molecules to spark evolution.
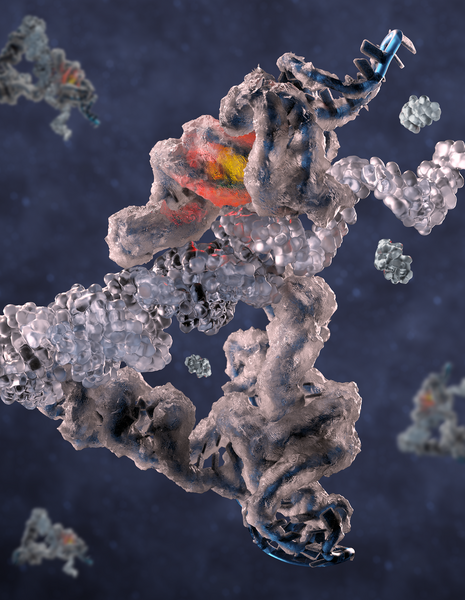 The picture shows an RNA polymerase ribozyme thought to be implicated in the origin of life. The ribozyme is shown frozen in ice to symbolize how it was frozen in time for imaging and how it works best under ice-cold conditions. A yellow/red light highlights the active site, and the proposed position of the template-product helix is shown in transparent. Image Credit: Rune Kidmose.
The picture shows an RNA polymerase ribozyme thought to be implicated in the origin of life. The ribozyme is shown frozen in ice to symbolize how it was frozen in time for imaging and how it works best under ice-cold conditions. A yellow/red light highlights the active site, and the proposed position of the template-product helix is shown in transparent. Image Credit: Rune Kidmose.
But more effective protein machines seem to have replaced the antiquated replicase in modern biology, as though it has vanished into the mists of time. Scientists have been working to replicate an analog of RNA replicase in the lab to bolster the RNA world theory. Even though these molecular "Doppelgangers" of the ancient replicase have been found, it has been difficult to ascertain their precise molecular structure and mode of action.
Structure of an Ice-Loving RNA Replicase
Using cryogenic electron microscopy (cryo-EM), a group of researchers has now reported the first atomic structure of an RNA replicase in a study published in PNAS. The Holliger group designed the RNA replicase under study to be effective at duplicating long templates using nucleotide triplets in the eutectic ice phase (slush-ice-like).
Image Credit: Kateryna Kon/Shutterstock.com
Emil L. Kristoffersen, an Assistant Professor at Aarhus University, Denmark, returned from postdoctoral research at the Holliger lab and arranged a cooperation with the Andersen lab to ascertain the structure of the RNA replicase by cryo-EM.
The structure has a remarkable resemblance to protein-based polymerases, featuring domains arrayed in a molecular form that resembles an open hand for substrate discrimination, polymerization, and template binding.
It was surprising to find that a ribozyme that we evolved artificially in the test tube would display features of naturally occurring protein polymerases. This indicates that evolution can discover convergent molecular solutions no matter if the material is RNA or protein."
Philipp Holliger, Program Leader, MRC LMB Cambridge, United Kingdom
Model for RNA Synthesis in an RNA World
The researchers conducted a thorough mutational analysis to identify the critical RNA structural components to gain a better understanding of how the RNA replicase functions.
In addition to confirming characteristics of the catalytic site, this analysis also demonstrated the significance of two kissing-loop interactions that bind the catalytic subunits and scaffolding together and the significance of a particular RNA domain for fidelity, or the precision with which the replicase copies RNA strands.
Even if the structure of the replicase "in-action" during active RNA copying could not be ascertained, a model for RNA-based RNA copying that is in line with all experimental evidence could be constructed.
Cryo-EM is a powerful method for studying the structure and dynamical features of RNA molecules. By combing cryo-EM data with experiments, we were able to build a model of the inner workings of this complex RNA machine."
Ewan McRae, Houston Methodist Research Institute
Ewan McRae also did a postdoc in cryo-EM in the Andersen lab at Aarhus University.
Inspiration for RNA Nanotechnology and Medicine
An intriguing initial look at an RNA replicase that is believed to be at the base of the tree of life is given by this study. However, compared to protein-based polymerases, the RNA-based replicases that are now under development are incredibly inefficient and are unable to support their own replication and evolution.
The structural knowledge of the presented study may contribute to the development of more effective replication methods, bringing them one step closer to creating RNA world situations in a test tube.
The properties of RNA replicases may be further improved by using chemical modifications that could exist in an RNA world. In addition, research into the origin of life leads to the discovery of several novel RNA building blocks that may be used in the emerging field of RNA nanotechnology and medicine.”
Ebbe Sloth Andersen, Associate Professor, Aarhus University
Source:
Journal reference:
McRae, E. K. S., et.al. (2024). Cryo-EM structure and functional landscape of an RNA polymerase ribozyme. Proceedings of the National Academy of Sciences (PNAS). doi.org/10.1073/pnas.2313332121.