Changes in gene expression in sebaceous glands have been spatially mapped for the first time in a collaborative project between Leipzig University's Interdisciplinary Centre for Bioinformatics (IZBI) and the Faculty of Veterinary Medicine. The study finds novel candidates for modulating sebum production and documents high-resolution changes in gene expression during sebum synthesis. The researchers published the results in the esteemed Journal of Biological Chemistry.
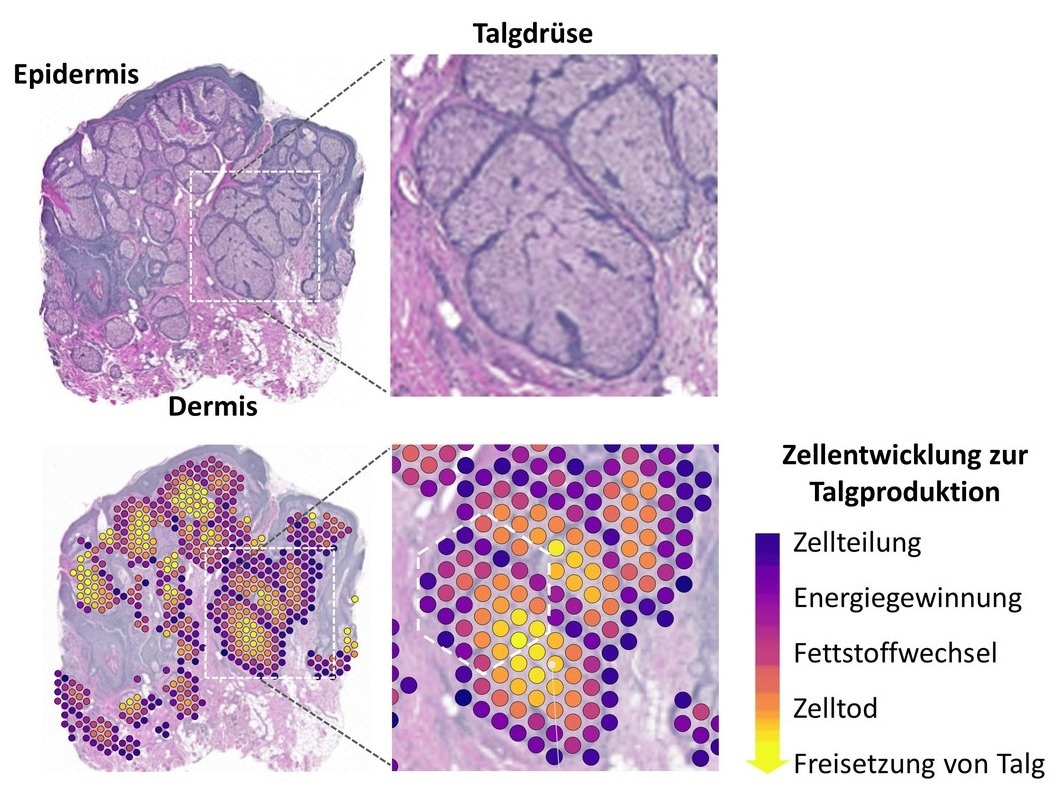 Histological structure of the sebaceous glands examined (haematoxylin-eosin stain). The area in the white rectangle is enlarged on the right. The clusters generated from the molecular biology and bioinformatics analysis (colored circles; left and center) represent the specific gene expression in this area. They were transferred to the original tissue and demonstrate the continuous development of the sebaceous gland cells through consecutive modulation of cellular functions (right). Image Credit: Interdisciplinary Centre for Bioinformatics (IZBI) at Leipzig University
Histological structure of the sebaceous glands examined (haematoxylin-eosin stain). The area in the white rectangle is enlarged on the right. The clusters generated from the molecular biology and bioinformatics analysis (colored circles; left and center) represent the specific gene expression in this area. They were transferred to the original tissue and demonstrate the continuous development of the sebaceous gland cells through consecutive modulation of cellular functions (right). Image Credit: Interdisciplinary Centre for Bioinformatics (IZBI) at Leipzig University
Sebaceous glands are necessary to keep the skin's structure and functionality intact. Holocrine secretion is the process by which cells in the gland's periphery gather a lot of lipid molecules, move them to the center of the gland, and then burst, releasing sebum, an oily secretion that lubricates and protects the skin's surface.
Other significant but mostly unidentified roles for sebum in humans and most other mammals include defense against UVB-induced apoptosis (cell death) and support of the skin's innate immune response. Sebum production may also impact the body's overall energy metabolism.
Acne and other skin disorders like psoriasis and eczema, which are chronic, itchy inflammations of the upper layers of the skin, are largely caused by sebaceous gland dysregulation. Furthermore, according to recent research, sebum production may impact the body's overall energy metabolism and have significant immunomodulatory effects. Hence, regulating sebum secretion could be a desirable therapeutic target for infections, metabolic disorders, and skin conditions.
Quite apart from their importance in skin health, sebaceous glands are an attractive and accessible model for understanding fundamental biological processes such as lipid synthesis, stem cell function or tumor development.”
Marlon R. Schneider, Study Supervisor, Professor and Head, Institute of Veterinary Physiology, Leipzig University
However, the alterations in gene expression that result in sebum production in sebocytes, or sebaceous gland cells, have not been thoroughly studied. In this recently published study, the two research groups map the progressive development of sebaceous gland cells up to cell lysis by integrating novel techniques like functional enrichment, pseudo-time analysis, and spatial analysis of gene expression.
The data demonstrates a sequential modulation of cellular functions that can be linked to particular gene sets, ranging from oxidative phosphorylation and cell proliferation to lipid synthesis and cell death. These stages of cell differentiation are based on four stages that were generated by unsupervised clustering.
The method is highly data-driven. Researchers around the world can use the tool to search and independently analyze our data.”
PD Dr. Hans Binder, Former Director, Interdisciplinary Centre for Bioinformatics, Leipzig University
Binder emphasizes the importance of an online bioinformatics tool developed in parallel.
Combining various approaches can result in a thorough and novel understanding of the gene expression dynamics underpinning sebaceous gland differentiation. The information points to the ongoing development of sebaceous cells through a series of cellular function modulations associated with particular gene sets.
The study points to future experimental approaches with medical relevance, identifies new players in sebum production, and offers new ways to modulate sebum production.
Next target: Skin Conditions and Tears
The next stage will involve a more thorough characterization of the sebaceous gland's altered gene expression regarding specific skin conditions, such as psoriasis, eczema, and tumors of the sebaceous glands. The current research endeavors will employ the subsequent iteration of the spatial transcriptome analysis technique, which facilitates the acquisition of single-cell-level outcomes.
In 2020, the prestigious journal Nature named spatially resolved gene expression its Method of the Year. As a combination of molecular biology and microscopy, it is highly data-driven and, in combination with sophisticated bioinformatics, opens up new insights into the organization of life processes in space and time at cellular resolution.”
Dr. Maria Schmidt, Study First Author, Leipzig University
Source:
Journal reference:
Schmidt, M., et al. (2024) A spatial portrait of the human sebaceous gland transcriptional program. Science Direct. doi.org/10.1016/j.jbc.2024.107442