From a genetic perspective, the formation of an R-loop is a critical risk for bacteria. An R-loop occurs when newly synthesized RNA binds back to its DNA template, creating a three-stranded structure. While R-loops play essential roles in certain cellular processes, their formation at the wrong time or location can be disastrous, causing DNA breaks, mutations, and even cell death.
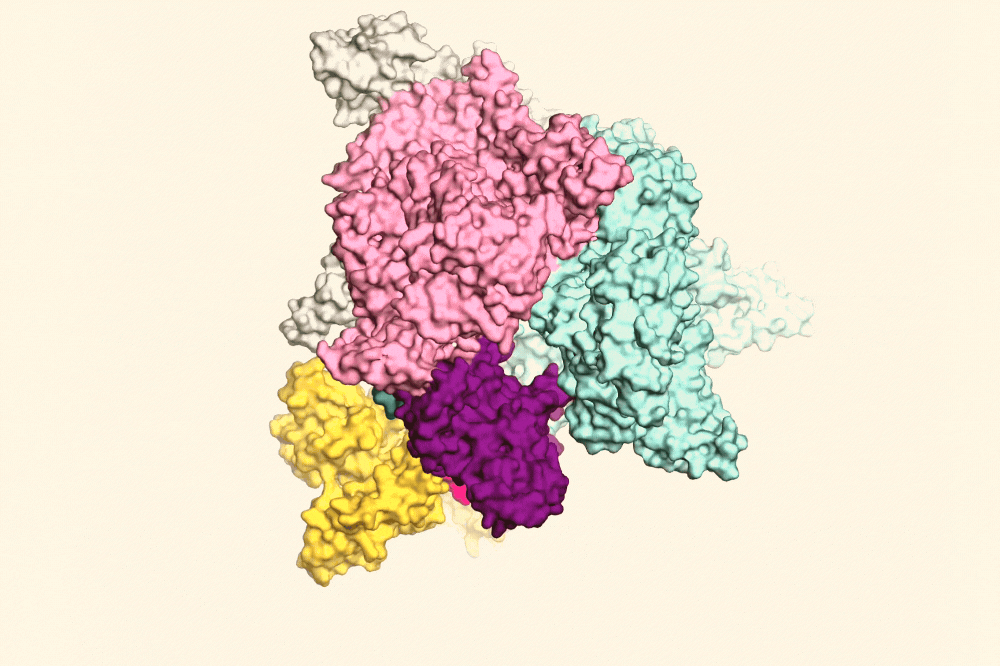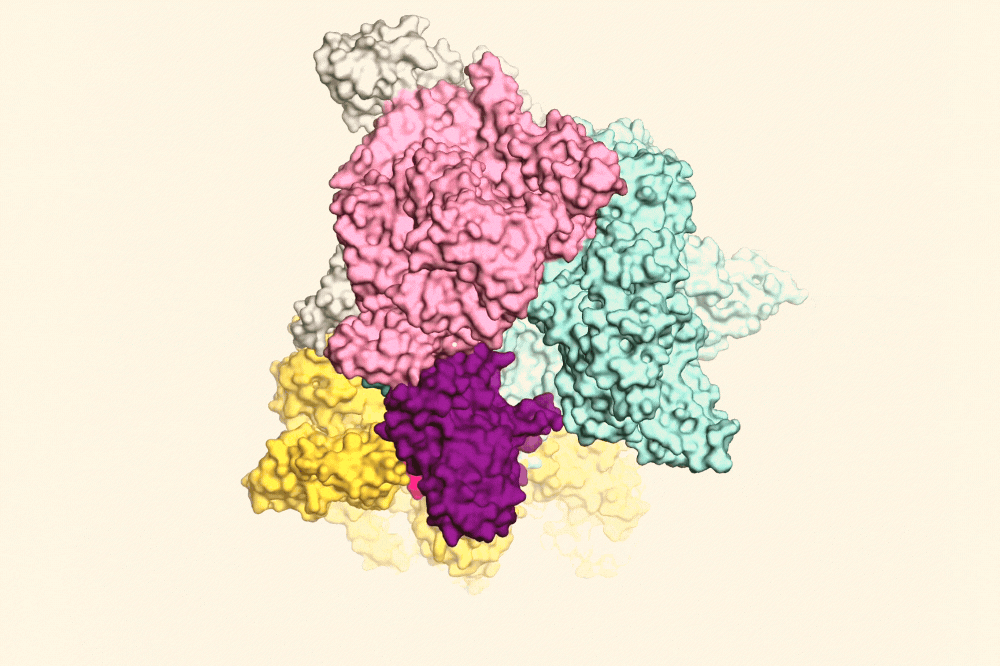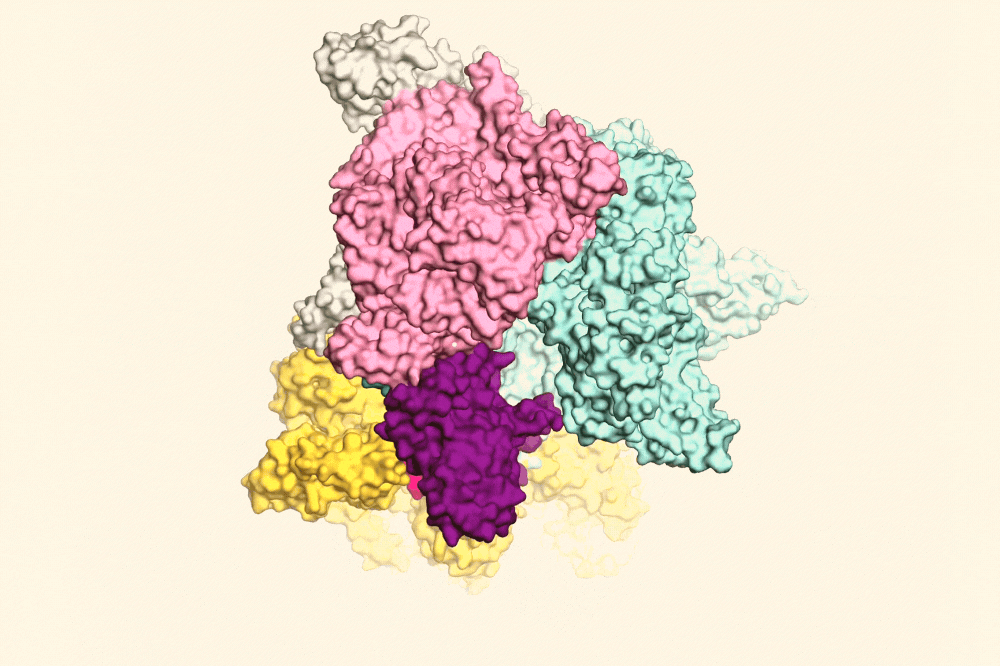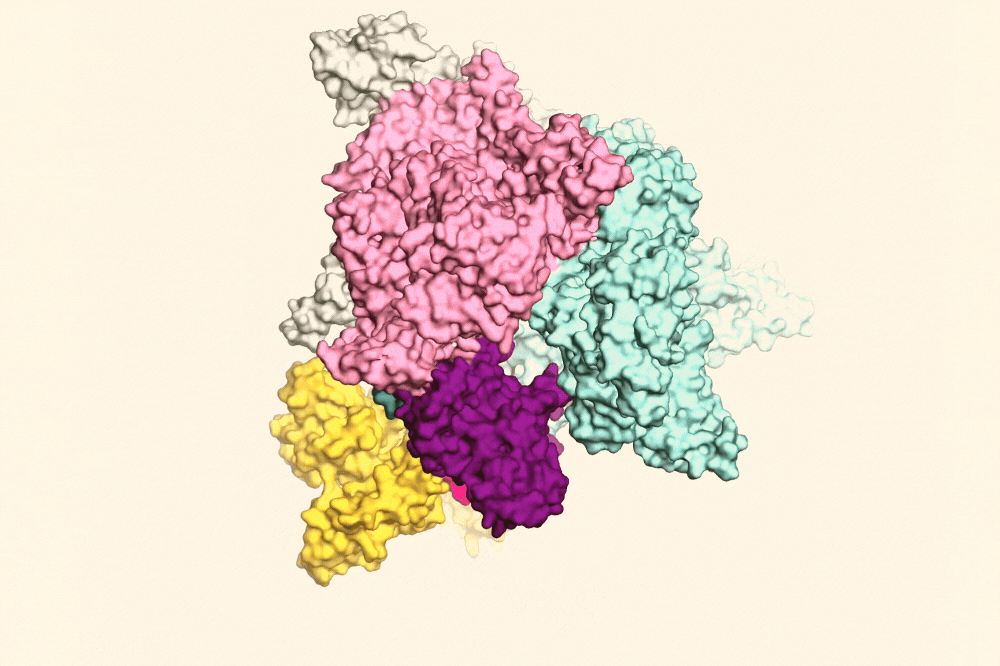 RapA (yellow) pulling open the RNAP clamp (purple) to help the RNAP fall off the DNA. Image Credit: The Rockefeller University
RapA (yellow) pulling open the RNAP clamp (purple) to help the RNAP fall off the DNA. Image Credit: The Rockefeller University
According to new research published in the journal Nature Structural & Molecular Biology, the enzyme RapA stops R-loop formation in E. coli. This has significant ramifications for how all cells preserve genomic stability.
The results show that, without the action of a protein called RapA, the RNA polymerase (RNAP) enzyme, which is in charge of copying DNA into RNA, can produce a lot of R-loops in specific situations.
R-loops are generally bad news, so cells have many redundant mechanisms to prevent them from forming. We discovered that RapA, a protein that we have been curious about for years, is one of those key mechanisms.”
Seth Darst, Head, Laboratory of Molecular Biophysics, The Rockefeller University
The Jaws of Life
RNAP is essential for the transcription of DNA into RNA in all living organisms. Scientists have long known that transcription in bacteria starts when RNAP clamps onto a DNA strand and starts the process after getting the all-clear from sigma proteins.
However, it is still unclear exactly how transcription ends. For example, recent research has revealed that RNAP frequently stays clamped to the DNA even after the freshly completed RNA transcript is released; however, it was not well understood why and how this occurs.
The Darst lab had identified RapA, an ATPase that obviously interacted with RNAP but had no apparent purpose, back in the 1990s.
“At the time, we really could not figure out what RapA was doing,” he said.
However, Darst's interest in the enigmatic protein was rekindled a few decades later when a different research team discovered that E. coli exposed to stressful, high-salt conditions could not grow without RapA.
His group started analyzing how RapA interacts with RNAP and how it stays clamped to DNA after transcription termination using cryo-EM. In contrast to the linear DNA frequently used in structural studies, they selected negatively supercoiled DNA, an underwound and twisted form of the double helix, because it more closely resembles the natural state of bacterial DNA.
Ours is one of the first studies to use negatively supercoiled DNA in a cryo-EM experiment. This method helped us better visualize the topological state of the DNA, how the proteins rearrange themselves, and how they interact with the DNA.”
Joshua Brewer, Study First Author, The Rockefeller University
They discovered, to their surprise, that when RNAP is clamped to DNA after transcription is finished, it rarely sits idly. Alternatively, it can start transcription once more without the typical protection of sigma proteins. Without sigma, transcription initiation creates dangerous R-loops unless RapA can break the RNAP clamp first.
RNAP is like a big claw that closes around DNA. RapA binds RNAP and pulls the clamp open so that it falls off the DNA before it can create R-loops.”
Seth Darst, Head, Laboratory of Molecular Biophysics, The Rockefeller University
Beyond Bacteria
The role of RapA started to become more apparent. RNAP is more likely to stay clamped to DNA and form R-loops under specific conditions, as demonstrated by the genetic instability the team observed in bacteria engineered without RapA when exposed to stressful, high-salt conditions.
They also discovered that although Rho, a well-researched enzyme that can break down R-loops, is also present in E. coli, Rho is unable to compensate in the absence of RapA completely.
Brewer said, “When you knock out RapA, Rho has to work overtime. It appears that RapA and Rho work as complementary, not redundant, safeguards for genome stability when E. coli are subject to high-salt stress.”
The ramifications might be extensive. According to Darst, Brewer, and associates, RapA or a similar protein is an RNAP release factor, as it may be present in all bacteria and possibly all cells in addition to E. coli.
The discovery of comparable mechanisms in other organisms may spark new approaches to treating illnesses associated with transcription-related genome instability.
Darst said, “We predict that other enzymes likely serve similar functions across the tree of life. The more we learn about these mechanisms, the more we deepen our understanding of how cells safeguard their genomes.”
Source:
Journal reference:
Brewer, J. J., et al. (2025) RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation. Nature Structural & Molecular Biology. doi.org/10.1038/s41594-024-01447-8.