The interaction between antigens and antibodies, also called immunoglobulins, underpins the immune response. Antigens are typically exogenous and endogenous molecules that can engage the body’s immune system.
Antigens include metabolites or products produced from cellular processes to a host of molecules that enter the body by ingestion, injection, or inhalation. Antibodies are products of the B lymphocytes, that differentiate into plasma B cells in order to mass-produce antibodies. B cells are responsible for ensuring antibody recognition and are structurally matched to the antigen.
Immunology - Introduction to Antibodies
Antibody structure
The antibody is comprised of many small protein units or domains characterized by two copies of a structural motif called β-sheets. These β-sheets are arranged in an anti-parallel fashion to form a three-dimensional structure called the immunoglobulin (Ig) fold of the Ig superfamily. This fold is comprised of 7 or more strands, arranged as two sheets to form a 3D structure called a domain that is 110 amino acids in length.
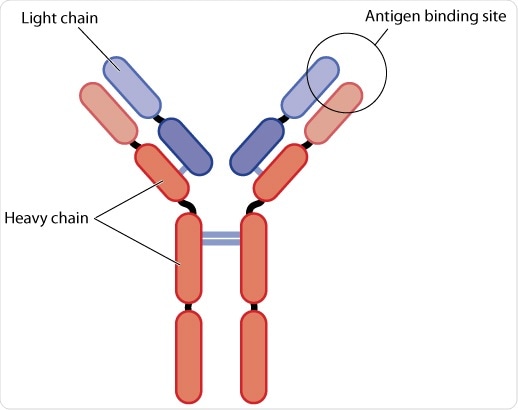
Heavy and light chains
A combination of four domains forms a heavy chain (H) and a combination of two domains form a light chain (L). Domains can be classified into constant (C) or variable (V). The convention for naming these domains is to denote the chain (H or L) in superscript. Two identical copies of both the light and heavy chains form a ‘Y’ shaped molecule.
Variable and constant regions
The heavy chain is comprised of one variable (VH) at the amino (N-)terminus (tip), and three constant domains CH1, CH2, and CH3. CH3 is present at the carboxy (C-)terminus (tail). The light chains are comprised of one variable (VL) and one constant (CL) domain. Within this basic H2L2 structure there are two major regions: Fc (‘fragment, crystallizable’) and Fabs (fragment, antigen-binding).
The Fab domain, which determines recognition and binding of the antigen, and Fc controls how it engages with the receptors on cells of the immune system or immune proteins to clear the antigen. The Fc and Fab are connected by a hinge region whose angles may vary, allowing the molecule to effectively bind antigen.
Schematic diagram of an antibody. The antibody is comprised of 2 heavy chains and two light chains. Both are comprised of a variable (V) and a constant domain (C). The amino-terminal domains in red correspond to the region of antigen binding, whilst the remained of the antibody domains exact effector functions. (Blamb/Shutterstock)
The central paradigm of antibody recognition
Immunoglobulins can be classified according to the type of heavy chain they contain and, in some cases, in their subunit structure. Although all IgG molecules have the same overall structure, they differ in the antigen they recognize. This property is called hypervariability.
The N-terminal region is called the Fv and encompasses the outer VH and VL of the antibody; a crevice between the two is the antigen-binding site. These regions are also referred to as complementarity-determining regions (CDRs) as they determine the shape of the binding site.
When an antibody binds to an antigen, some of the surface areas on the antibody molecule is buried by the antigen; this decreases with the size of the antigen as less of the antibody surface is required for binding.
Antibody maturation
The surface of the antibody that recognizes the antigen is the product of the genetic diversity encoded by the genes of the immune system. Yet, the number of antibody genes is far smaller than the observed diversity. Rather, diversity is generated by recombination and imprecise joining of antibody gene segments. These genes produce the molecular surface formed by the CDRs, which form the site of antigen recognition.
The V, D, and J segments encode the heavy chain, whilst the V, C, and J segments encode the light chain. There are many V, D, J, and C segments and so their combinatorial possibilities are vast. The antibodies that are produced are of low affinity and specificity; for an antibody to optimally function, it must undergo a process called maturation. Somatic hypermutation
A series of random point mutations in the V region arise by somatic hypermutation, through which binding specificity and affinities of antibodies are altered.
In the second process, specific clones of B cells are selected by specialized cells called the follicular dendritic cells (FDCs) at specific sites called Germinal Centres in the lymph nodes and spleen. The B cells that produce the antibodies with the highest affinity and specificity outcompete other clones; this is called positive selection. This positive selection is augmented by survival signals provided by specialized immune cells called Tfh cells of the germinal center.
Monoclonal and polyclonal antibodies
Antibodies can be monoclonal or polyclonal. Monoclonal antibodies recognize only one type of epitope present on an antigen and are produced by identical B-cells. By contrast, polyclonal antibodies can bind to multiple epitopes on an antigen; they arise from different B cells whose V, D, J, and C segments are varied. The latter is therefore highly variable and therefore tolerate minor changes in the antigen.
Antibody-antigen interactions
Antibody-antigen interactions underpin the immune response. The diversity in antibodies necessary for successful clearance of the offending organism or allergen arises from gene rearrangement and hypermutation in the structural regions that form the site of antigen recognition.
Further Reading