Reviewed by Danielle Ellis, B.Sc.Jan 22 2024
The world’s first deep generative model for RNA design was created through a joint research project between Professors Hirohide Saito (Department of Life Science Frontiers, CiRA, Kyoto University) and Michiaki Hamada (Waseda University).
 Deep generative model of functional RNA. Image Credit: CiRA, Kyoto University
Deep generative model of functional RNA. Image Credit: CiRA, Kyoto University
Although aptamer and antisense oligonucleotide medications have been available since the 2000s, the public did not become aware of RNA-based therapeutics until the creation of SARS-CoV2 mRNA vaccines, which were used to combat the COVID-19 pandemic.

Image Credit: CROCOTHERY/Shutterstock.com
On the other hand, due to its enormous potential for medical applications as well as basic biological research and biotechnology, RNA engineering has been at the forefront of science for many years. Therefore, there is a great deal of interest in transforming the methods used now to design RNA sequences.
Surprisingly, there is still no flexible computing environment available for the design of functional RNA. The majority of current methods, like CRISPR gRNA, mRNA, or particular riboswitches, work by reconstructing particular secondary structures or are limited to specific sequence types.
The accuracy of these conventional methods is inevitably limited by structural prediction and optimization algorithms since they usually rely on RNA secondary structure prediction and optimization. Therefore, a new strategy was required to get around these restrictions and create strong and effective computational techniques for creating RNA with the desired functions.
By concentrating on RNA families, which are sequence groups with thousands of functional RNAs endowed with identical functions, the research team hoped to avoid these issues. Multiple sequence alignment can produce a consensus secondary structure from which new sequences can be generated, even with just a few hundred sequences.
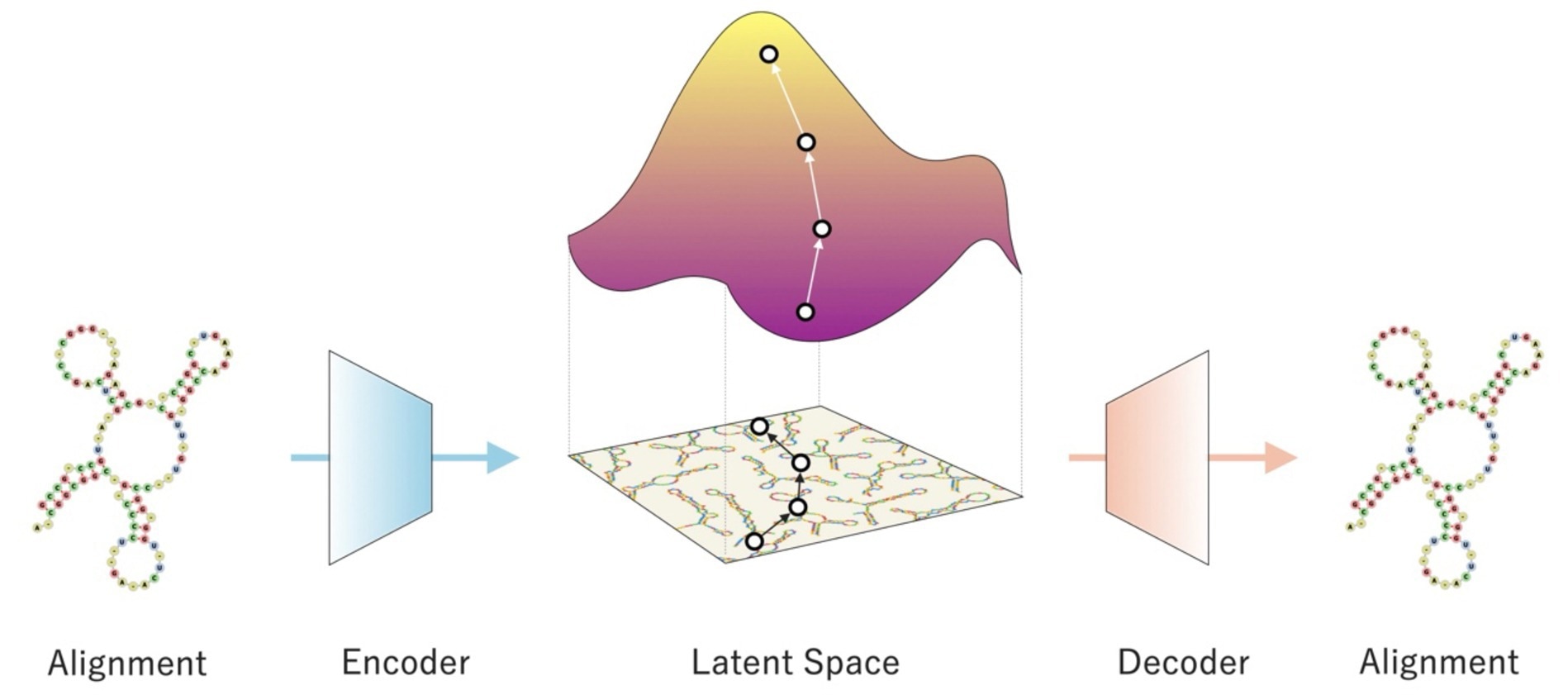
The researchers called their deep generative model the RNA family sequence Generator, or RfamGen, which is the first deep generative model for functional RNA design in the world because it theoretically works with any functional RNA family.
RfamGen combines two methods: variational autoencoder and covariance model. A kind of statistical framework for consensus secondary structure and RNA alignment that measures sequence and structural variations quantitatively is the covariance model.
To lessen the complexity involved in examining the exponentially large sequence space for RNA sequence optimization, the variational autoencoder is a deep generative model with an internal representation known as “latent space.”
By combining these two ideas, the researchers created a system that, for the first time, can logically explore novel RNA designs by learning sequence and structural information.
Initially, the group contrasted RfamGen, which takes into account alignment as well as secondary structural data, with models that take into account either alignment, secondary structural data, or neither.
RfamGen demonstrated a markedly improved capacity to produce high-quality RNA sequences for the 18 RNA families that were tested (each with alignments consisting of at least 10,000 sequences). In addition, the researchers evaluated RfamGen’s performance when it was given a restricted set of input sequences to learn from. RfamGen effectively produced RNA sequences with high scores despite having only been trained on 500 input sequences, proving its effective generative capacity.
Subsequently, the researchers trained RfamGen on a total of 629 RNA families, each containing at least 100 sequences from the Rfam database, and discovered that RfamGen outperforms other systems by a significant margin. Additionally, the researchers randomly synthesized multiple RNA sequences produced from training it with a variety of self-cleavage ribozymes and from randomly sampling a covariance model in order to assess how well-generated RNA sequences function.
Interestingly, RfamGen-generated sequences exhibited enzymatic activity, whereas randomly sampled sequences did not. This suggests that RfamGen picked up crucial functionality-related features from the training data.
Finally, the researchers benchmarked RfamGen-generated sequences against naturally occurring glmS sequences using the ligand-dependent self-cleavage activity of the glmS ribozyme. Approximately 500 natural glmS ribozyme sequences were used to train RfamGen, and 1,000 generated sequences were obtained by sampling the “latent space.”
The scientists tested these 1,000 generated sequences, 761 natural sequences in the glmS ribozyme family (RF00234), and 100 sequences with kinetic measurements from an earlier report using a massively parallel assay.
The team discovered that the generated sequences had higher cleavage rates than the natural sequences, indicating that RfamGen is successful in producing high-quality sequences that are either as efficient or more efficient than some natural sequences. In addition, the generated sequences were observed to have a similar distribution of cleavage kinetics as the natural sequences.
The era of RNA-based bioengineering is about to enter its golden age. The research team believes that RfamGen will be a fundamental driving force to propel RNA biology into a new era and enable RNA-based discoveries and applications by building this deep generative model for functional RNA design.
Source:
Journal reference:
Sumi, S., et al. (2024) Deep generative design of RNA family sequences. Nature Methods. doi.org/10.1038/s41592-023-02148-8.