The genes that are activated in a single cell can now be determined thanks to advances in high-throughput biological studies. However, interpreting the resulting complex datasets might be difficult. CAPITAL, a novel computational tool for comparing complex datasets from single cells, has now been developed by a group at Osaka University.
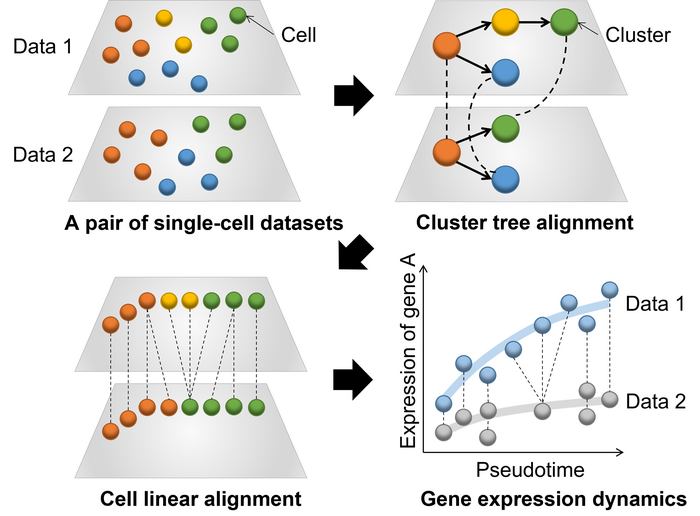 Overview of CAPITAL: an algorithm for comparing pseudotime trajectories with branches. Image Credit: Yuki Kato
Overview of CAPITAL: an algorithm for comparing pseudotime trajectories with branches. Image Credit: Yuki Kato
RNA sequencing reveals a fraction of the whole population of genes that are actively expressed, or “switched on.” With the advancement of technology, it is now possible to sequence the RNA population of a single cell.
Since each cell can be explicitly examined instead of all of the different cell types being pooled, this can yield a wealth of information on the precise gene expression changes associated when a huge population of mixed cells goes through dynamic, transitional processes, like differentiation or cell death.
CAPITAL was created primarily to compare large datasets from individual cells going through transitional stages. The “pseudotime trajectory” used in these investigations places the cells along a hypothetical path that represents how they move during the transitional phase.
These trajectories can be quite complex and branching; they are not always simple and linear. Prior to the group’s breakthrough, only linear trajectories could be reliably and automatically aligned for comparison. Now, however, complicated branching trajectories may also be aligned and compared.
They evaluated the algorithm used for CAPITAL, which employs a technique known as tree alignment, on both fabricated datasets and real datasets derived from bone marrow cells. The findings showed significant improvements over the previous computational algorithms, proving that CAPITAL is statistically more precise and trustworthy.
A strong technique known as trajectory comparison can, for instance, determine the dynamics of gene expression in many species to shed light on evolutionary processes.
We showed in this study that CAPITAL can reveal the existence of different molecular patterns between humans and mice even when the expression patterns are similar and appear to be conserved. This will allow the identification of novel regulators that determine cell fates.”
Reiichi Sugihara, Study Lead Author, Osaka University
This technology is not just applicable to this kind of data.
Our novel computational tool can be applied to a wide range of high-throughput datasets, including pseudotemporal, spatial, and epigenetic data.”
Yuki Kato, Study Senior Author, Osaka University
By comparing single-cell trajectories globally, this potent new tool will enable the discovery of novel disease-associated genes that were previously undetectable by comparative methods. As a result, CAPITAL marks a noteworthy development in the discipline of single-cell biology.
Source:
Journal reference:
Sugihara, R., et al. (2022) Alignment of single-cell trajectory trees with CAPITAL. Nature Communications. doi.org/10.1038/s41467-022-33681-3.